A high-content screening assay for small-molecule inhibitors of the Colony Stimulating factor 1 Receptor (CSF1R) signaling pathway
2 Beijing Institute of Pharmacology and Toxicology, Beijing 100850, China, Email: weichen@northwestern.edu
Received: 19-Aug-2017 Accepted Date: Sep 22, 2017; Published: 26-Sep-2017
Citation: Yang S, Chen W, Long L, et al.. A high-content screening assay for smallmolecule inhibitors of the Colony Stimulating factor 1 receptor (CSF1R) signaling pathway J Pharmacol Res September-2017;1(1):10-16.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
The aim of the study was to establish a rapid and reliable method for screening CSF1R (Colony Stimulating factor 1 Receptor) signaling pathway inhibitors. Plasmid containing human CSF1R gene was constructed and transfected into U2OS cells stably expressing GFP-STAT1 (green fluorescent protein-signal transducers and activators of transcription) fusion protein. A cell line stably expressing both CSF1R and GFP-STAT1 fusion protein was generated, and named CSF1R/GFPSTAT1_U2OS. The CSF1R was expressed and confirmed to be functional, as exposure of rhM-CSF to the cells induce significant GFP-STAT1 nuclear translocation. The screening protocol for CSF1R inhibitors was then optimized basin on this. According to the results, 30 min of rhM-CSF exposure induce maximum protein nuclear translocation, and the EC50 value for rhM-CSF to induce GFP-STAT1 nuclear translocation was 43.50 ± 3.68 ng/mL, and the maximum STAT1 nuclear translocation was induced by about 300 ng/mL of rhM-CSF. The mean z’ factor of the assay was 0.77, confirming the robustness and reliability of current method. Three known CSF1R inhibitors, GW2580, Sunitinib or Imatinib mesylate concentration dependently blocked the rhM-CSF induced GFP-STAT1 nuclear translocation. Using the established assay, 81 compounds were screened, and 4 compounds emerge as positive hits. In conclusion, a U2OS cell line stably expressing CSF1R and GFP-STAT1 was generated, which can be used in screening and evaluating of novel CSF1R signaling pathway inhibitors by simply monitoring the nuclear translocation behavior of GFP-STAT1.
Keywords
CSF1R; Tyrosine kinase; High content screening; STAT1; GFP
Introduction
Colony stimulating factor 1 receptor (CSF1R), also known as M-CSFR, c- Fms or CD115), a receptor tyrosine kinase (RTK), plays important roles in survival, proliferation and differentiation of the monocyte and macrophage [1]. As a tyrosine kinase, activation of CSF1R by CSF-1 or Interleukin-34 (IL-34) binding [2,3] can lead to activation of several downstream signaling pathways include PI3K/Akt pathway [4], MAPK/ERK pathway [5], and PLC pathway [6], as well as the activation of STAT (signal transducers and activators of transcription) family transcription factors [7]. Mutation of CSF1R or aberrant expression of CSF1 or CSF1R has been observed in ovarian, breast, prostate cancers, as well as several blood malignances, and also had been proved to promote cancer proliferation, invasion and metastasis [8]. In fact, the gene responsible for CSF1R is also recognized as c-fms proto oncogene [9]. Besides, CSF1R signaling pathway is also involved in inflammatory diseases and bone diseases.
Blockade of the CSF1R signaling axis has been regarded as an effective way in controlling the above mentioned diseases [10]. Thus the research and development of CSFR pathway inhibitors, especially CSF1R tyrosine kinase inhibitors, faces unmet needs [8]. Some selective CSF1R inhibitors such as GW2580 and Ki20227 have recently been discovered and confirmed to provide benefits in rheumatoid arthritis [11,12]. Additionally, some other RTKs inhibitors like Sunitinib, Imatinib mesylate and sorafenib also have been found to be CSF1R inhibitors and have effective therapeutics against the autoimmune demyelinating disease [13-15]. Regardless of the effectiveness in controlling these diseases, satisfying CSF1R signaling pathway inhibitors remains lacking, partly due to the lack of reliable and efficient screening method during the early stages of CSF1R inhibitors development. The most widely used assays in screening of CSF1R inhibitors include ATP site-dependent competition binding assays [16], cell proliferation assays [11] and immunoblotting assays [17]. Additionally, some commercialized kits based on Fluorescence Polarization, Fluorescence Resonance Energy Transfer and horseradish peroxidase coloration technologies have also been developed for the discovery of RTKs inhibitors including CSF1R inhibitors. However, all these methods listed above have shortcomings at either accuracy or efficiency, i.e. throughput.
In current study, we established a novel screening assay for CSF1R signaling pathway inhibitors based on High Content Screening [HCS] platform, which represent a reliable and efficient way of inhibitor screening. The establishment of the assay was based on the wellrecognized fact that CSF1R activation by CSF1 or IL34 will result in activation and nuclear translocation of STAT1, who is a transcription factor downstream of several cytokines and growth factors. CSF1R activation and phosphorylation results in activation of JAK kinases include TYK2 and JAK1, which then phosphorylate and activate STAT17, phosphorylation of STAT1 by CSF1R can also be achieved through SRC kinase (Figure 1A) [18,19]. Activated STAT1 then forms heterodimer and translocate into the nuclear to drive expression of related genes [20]. Based on this, a cell line stably expressing human CSF1R and GFP labeled STAT1 was generated, and then the efficacy of the cell line in screening of CSF1R inhibitors was further validated. By monitoring the CSF induced nuclear translocation of GFP-STAT1 after compounds treatment; the assay provides a reliable and efficient way of screening CSF1R signaling pathway inhibitors.
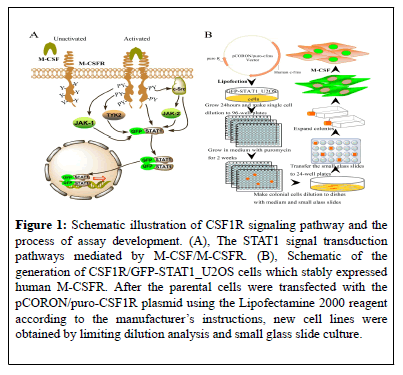
Figure 1: Schematic illustration of CSF1R signaling pathway and the process of assay development. (A), The STAT1 signal transduction pathways mediated by M-CSF/M-CSFR. (B), Schematic of the generation of CSF1R/GFP-STAT1_U2OS cells which stably expressed human M-CSFR. After the parental cells were transfected with the pCORON/puro-CSF1R plasmid using the Lipofectamine 2000 reagent according to the manufacturer’s instructions, new cell lines were obtained by limiting dilution analysis and small glass slide culture.
Materials and Methods
Compounds
GW2580, sunitinib (also known as sutent), and Imatinib mesylate were synthesized in the new drug design center of our institute as described previously [21-23]. The structure of the compounds was validated by NMR and MS, and their purity were ≥ 98% as detected by HPLC analysis. The 81 chemical entities were also designed and synthesized in our institute.
Reagents
Hoechst 33342 was purchased from Invitrogen (NY, USA). Neomycin (G418) and puromycin were obtained from Merck (Darmstadt, Germany). Recombinant human macrophage colony-stimulating factor (rhM-CSF) was purchased from Peprotech (NJ, USA).
Cell culture
Human osteosarcoma (U2OS) cells stably expressing GFP-STAT1 fusion protein (GFP-STAT1_U2OS cells) were obtained from GE Health (GE, USA) and were cultured in DMEM supplemented with 10% FBS, and 500 μg/ml G418. The culture medium for the newly generated CSF1R/GFPSTAT1_ U2OS cells were DMEM supplied with 10% FBS, 500 μg/ml G418 and 2 μg/ml puromycin. M-NFS-60 cells were cultured in RPMI-1640 medium supplemented with 7.5% FBS, 2.5% horse serum, and 10 ng/mL rhM-CSF. The assay medium was DMEM supplied with 10 mM HEPES and 0.2% BSA.
Plasmids construction
The full-length human CSF1R gene cDNA fragment was released with BamH1 from pSM-CSF1R plasmid (a kind gift from Dr. C.J. Sherr and Dr. Martine F. Roussel), and cloned into PUC18 vector (TaKaRa, Dalian, China) to obtain PUC18-CSF1R (R). The inserted fragment was released with EcoR1 and Xba1 from PUC18-CSF1R (R) and recloned into pCORON/puro to obtain pCORON/puro-CSF1R plasmid.
Generation of CSF1R /GFP-STAT1_U2OS cells
The flow sheet of generating CSF1R /GFP-STAT1_U2OS cell line is showed in Figure 1B. Basically, GFP-STAT1_U2OS cells was transfected with pCORON/puro-CSF1R plasmids. The transfected cells were diluted and cultured in 96-well plates at the density of 1 cell/well, and exposed to G418 and puromycin continuously. Then the formed single clones were individually transferred into dishes with small glass sliders. 10 days later, the small glass sliders on which a new single cloning cell was grown were transferred to 24-well plates. Positive clones was defined as cells exhibited apparent nuclear translocation after stimulation with rhM-CSF. Positive clones were further validated and the best one was selected and named as CSF1R /GFP-STAT1_U2OS.
Immunofluorescence Assay
Immunofluorescence assay was used to examine the expression status of CSF1R in the newly generated cell lines. Basically, cells cultured in 96 well plate (Corning 3603) were fixed with 4% paraformaldehyde and permeabilized with 0.5% (v/v) Triton X-100 at room temperature. Cells were then exposed to primary antibody against human CSF1R (Santa Cruz, USA) for 1 hour, followed by PBS rinse thrice and exposure to Alexa Flour 546 conjugated secondary antibody (anti-rat IgG, Invitrogen, USA) for another 1 hours. Finally, the cells were counterstained with Hoechst 33342 and fluorescent images were captured under the IN Cell Analyzer 1000 (GE, USA) by channel 1 (Ex/Em=360 nm/460 nm), channel 2 (Ex/Em=475 nm/535 nm) and channel 3 (Ex/Em=535 nm /620 nm).
Nuclear Translocation Assay
Cells were seeded in the 96-well plates (Corning 3603) at a density of 1 × 104 cells/well in 100 μL cell culture medium and cultured overnight. The cells were washed twice with the assay medium and incubated in 37 for 15 minutes before 30 minutes of rhM-CSF stimulation. The cells were then fixed with 4% (v/v) formaldehyde for 20 minutes and stained with 1μM Hoechst 33342 for 30 minutes. After washing twice with PBS, fluorescent images were acquired by the IN Cell Analyzer 1000 (GE Healthcare, NJ, USA; 20 × objective) through channel 1 (Ex/Em=360 nm/460 nm, 300 ms) and channel 2 (Ex/Em=475 nm /535 nm, 1000 ms). Images from five stochastic fields were obtained for each well. All images were analyzed using the Nuclear Trafficking Analysis Module of In Cell Analyzer 3.5 workstation (GE, USA). The translocation index, i.e. the ratio of total nuclear GFP fluorescence intensity to total cytoplasmic GFP fluorescence intensity for cell populations in five fields of each well, was used to describe the nuclear translocation of GFP-STAT1 from the cytoplasm to the nucleus.
For compounds screening, the cells were seeded and cultured overnight as described above. After washing twice with assay medium, cells were exposed to compounds in assay medium for 1 hour in cell culture incubator. After that, cells were stimulated with rhM-CSF and the following procedures were consistent with upward described.
Cell Proliferation Assay
The M-NFS-60 proliferation assay was carried out as described previously [11,24]. Briefly, M-NFS-60 cells were seeded into 96-wells plates at a density of 1 × 105 cells/well in 50 μL culture medium and then 50 μL/well of the medium containing compounds were added into each well. The cells were incubated for 72 h in CO2 incubator. After that, 10.0 μL of MTT (5.0 mg/mL) was added and incubated for another 4 hours. The culture medium was removed and 100.0 μL of 100% DMSO solution were added to each well to solubilize the formed formazan. The plates were read using the plate reader at 570 nm with reference at 630 nm wave length (Universal Microplate Reader EL800, BioTek, USA).
Statistical Analysis
Raw data were exported from the IN Cell Analyzer 1000 workstation 3.5 and all the data were presented as means ± S.D. Statistics were analyzed using One-Way ANOVA of SPSS 13.0. EC50 or IC50 values were generated using Origin 7.5. Z′ factor of the assay was calculated using the formula: Z′=1-3 (σp+σn)/|μp-μn|, where μp and σp are the mean and standard deviation of the positive control, and μn and σn are the mean and standard deviation of the negative control, respectively [25].
Results
Validation of the Established CSF1R/GFPSTAT1_ U2OS Cells
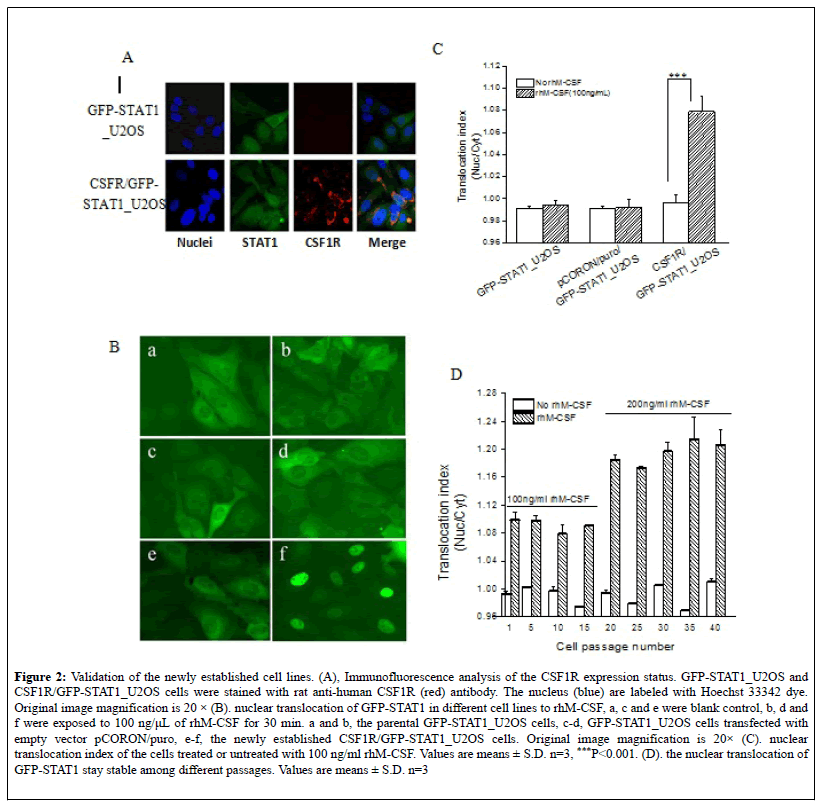
A schematic illustration of the underlying mechanism for the assay was available in Figure 1A. And the procedure of cell line establishment was described in detail in the Material and Method section, as well as in Figure 1B. The expression status of CSF1R in the newly generated cells was validated by immunofluorescence assay. As shown in Figure 2A, both the parental GFP-STAT1_U2OS cells and the CSF1R/GFP-STAT1_U2OS cells shows green fluorescence in the cytoplasm, while only the latter exhibit CSF1R fluorescence, confirming that the newly established cell lines express the protein of interest. In order to confirm the biological activity of human CSF1R in the positive clone cells, the cells were cultured overnight and treated with 100 ng/ml of rhM-CSF for 30 minutes. As shown in Figure 2B (f), GFP-STAT1 in CSF1R/GFP-STAT1_U2OS cells translocate into the nucleus from the cytoplasm, while the GFPSTAT1 in the parental cells or in the cells transfected with empty vector remain in the cytoplasm after rhM-CSF stimuli (Figure 2B, b and d). Quantitative analysis of fluorescent distribution revealed that rhM-CSF induce significant nuclear translocation of GFP-STAT1 in CSF1R/GFPSTAT1_ U2OS cells (Figure 2C). Additionally, the rhM-CSF induced GFP-STAT1 nuclear translocation remains stable during continuous cell passage, as the translocation index remains unchanged among different passages when cells were treaded with 100 ng/ml or 200 ng/ml rhM-CSF (Figure 2D). Together, these results indicated that human CSF1R was stably expressed in CSF1R/GFP-STAT1_U2OS cells and the expressed protein was functional.
Figure 2: Validation of the newly established cell lines. (A), Immunofluorescence analysis of the CSF1R expression status. GFP-STAT1_U2OS and CSF1R/GFP-STAT1_U2OS cells were stained with rat anti-human CSF1R (red) antibody. The nucleus (blue) are labeled with Hoechst 33342 dye. Original image magnification is 20 × (B). nuclear translocation of GFP-STAT1 in different cell lines to rhM-CSF, a, c and e were blank control, b, d and f were exposed to 100 ng/μL of rhM-CSF for 30 min. a and b, the parental GFP-STAT1_U2OS cells, c-d, GFP-STAT1_U2OS cells transfected with empty vector pCORON/puro, e-f, the newly established CSF1R/GFP-STAT1_U2OS cells. Original image magnification is 20× (C). nuclear translocation index of the cells treated or untreated with 100 ng/ml rhM-CSF. Values are means ± S.D. n=3, ***P<0.001. (D). the nuclear translocation of GFP-STAT1 stay stable among different passages. Values are means ± S.D. n=3
Optimization of High Content Screening (HCS) Assay for CSF1R Inhibitors
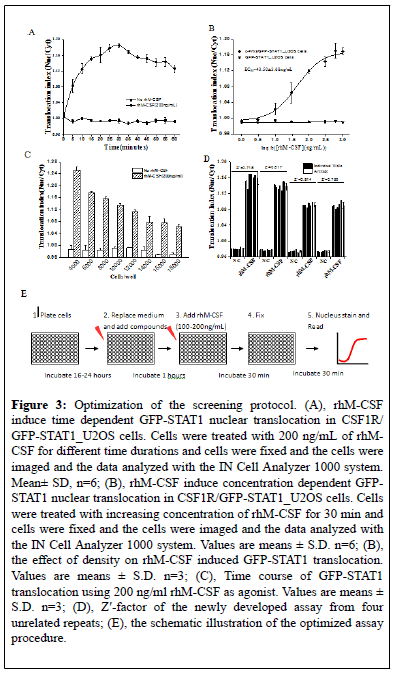
In order to make CSF1R /GFP-STAT1_U2OS cells to be more suitable for 96 well high content screening, we optimized the screening conditions. Firstly, CSF1R/GFP-STAT1_U2OS cells were exposed to 100 ng/mL of rhM-CSF for different time durations for up to 60 min at 5 min step. As shown in Figure 3A, rhM-CSF induce a time-dependent nuclear translocation of GFP-STAT1. Five minute of rhM-CSF was enough to induce significant protein nuclear translocation, while 30 min of stimuli induce maximum protein nuclear translocation. Then, cells were exposed to different concentration of rhM-CSF varying from 1-1000 ng/mL for 30 min, and a concentration dependent nuclear translocation of the GFPSTAT1 protein was observed (Figure 3B). The EC50 value for rhM-CSF to induced GFP-STAT1 nuclear translocation was 43.50 ± 3.68 ng/mL, and the minimum concentration for rhM-CSF to induce maximum STAT1 nuclear translocation was about 300 ng/mL Both the time dependent protein nuclear translocation and the concentration dependent protein nuclear translocation happens in CSF1R /GFP-STAT1_U2OS cells but not in GFP-STAT1_U2OS cells. We then optimized the seeding density of the assay by examining the nuclear translocation index under different seeding density. As shown in Figure 3C, the GFP-STAT1 nuclear translocation induced by 200 ng/ml rhM-CSF gradually decreased with the increasing density of cells. To guarantee the number of cells during image collection and data analysis, we suggest that 4000-5000 cells/well be the optimal seeding density in the form of 96 well screening. Considering the relative high price of cytokine, we suggest that 100-200 ng/mL of rhM-CSF be used during screening, 30 minutes as the optimal stimulation duration.
Figure 3: Optimization of the screening protocol. (A), rhM-CSF induce time dependent GFP-STAT1 nuclear translocation in CSF1R/ GFP-STAT1_U2OS cells. Cells were treated with 200 ng/mL of rhMCSF for different time durations and cells were fixed and the cells were imaged and the data analyzed with the IN Cell Analyzer 1000 system. Mean± SD, n=6; (B), rhM-CSF induce concentration dependent GFPSTAT1 nuclear translocation in CSF1R/GFP-STAT1_U2OS cells. Cells were treated with increasing concentration of rhM-CSF for 30 min and cells were fixed and the cells were imaged and the data analyzed with the IN Cell Analyzer 1000 system. Values are means ± S.D. n=6; (B), the effect of density on rhM-CSF induced GFP-STAT1 translocation. Values are means ± S.D. n=3; (C), Time course of GFP-STAT1 translocation using 200 ng/ml rhM-CSF as agonist. Values are means ± S.D. n=3; (D), Z'-factor of the newly developed assay from four unrelated repeats; (E), the schematic illustration of the optimized assay procedure.
The robustness of the established assay was then checked by four separate tests using the optimized conditions mentioned above. The Z’ factor of each test was calculated using the formula mentioned above. As shown in Figure 3D, Z′ factor of the HCS assays for CSF1R inhibitors from ranged from 0.72 to 0.82, with an average of 0.77 ± 0.049. Therefore, the established HCS assay were eligible, since Z′ factor between 0.50 and 1.00 were considered robust and reliable.
Validation of the Optimized HCS Assay for Compound Screening
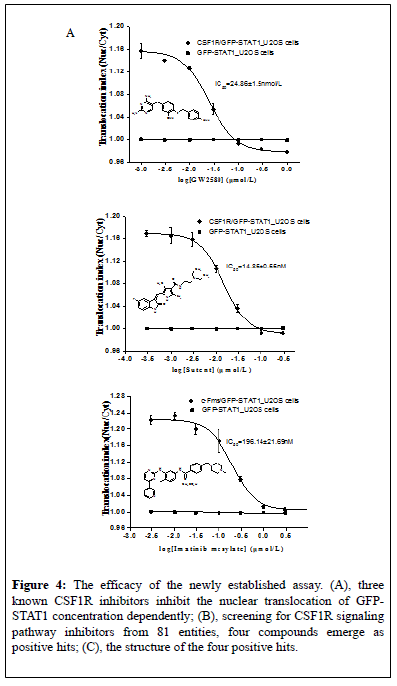
In order to further validate the reliability and practicality of the newly established HCS assay for CSF1R signaling pathway inhibitors, the inhibiting effects of three known CSF1R inhibitors, GW258011, sunitinib15, and imatinib14, were first evaluated by using this assay (Figure 3E). As shown in Figure 4A, all three compounds concentrationdependently inhibited GFP-STAT1 nuclear translocation induced by rhMCSF in CSF1R/GFP-STAT1_U2OS cells but not in GFP-STAT1_U2OS cells. The IC50 value was 24.86 ± 1.5 nM, 14.85 ± 0.55 nM and 196.14 ± 21.69 nM for GW2580, sunitinib and imatinib respectively. Obviously, Sunitinib and GW2580 were more potent CSF1R inhibitors than Imatinib. These results are consistent with that reported by IMPA biochemical method, NFS-60 cells proliferation assay and cellular pCSF1R and pERK assays [13]. The cytotoxicity of the compounds on CSF1R/GFPSTAT1_ U2OS cells in the screening assay were simultaneously evaluated by parameters include cell number, cell area and nuclear area. No significant influence on these parameters were observed (Figure 4B), indicating that the inhibition of three inhibitors on GFP-STAT1 nuclear translocation was not associated with cellular toxicity.
Figure 4: The efficacy of the newly established assay. (A), three known CSF1R inhibitors inhibit the nuclear translocation of GFPSTAT1 concentration dependently; (B), screening for CSF1R signaling pathway inhibitors from 81 entities, four compounds emerge as positive hits; (C), the structure of the four positive hits.
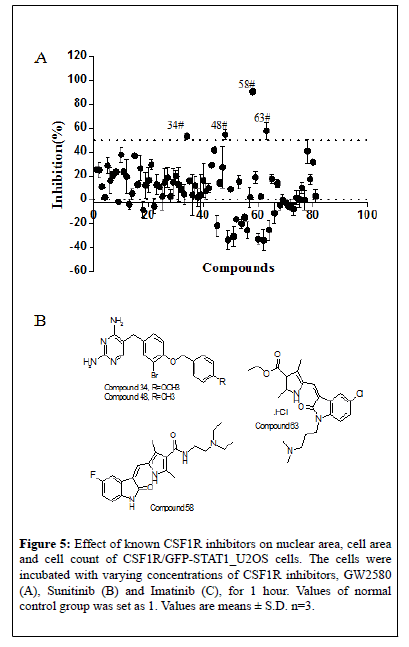
To further explore the potential of the established screening assay in identifying CSF1R signaling pathway inhibitors, 81 new chemical entities designed to target CSF1R were screened at a single concentration of 3 μM in duplicate plates using this assay. Among them, 4 compounds showed ≥ 50% inhibition of GFP-STAT1 nuclear translocation in the primary screen (Figure 5A). The 4 potential hits from the primary screen were then further tested in subsequent dose-response study. The IC50of the 4 compounds is shown in Table 1, and their chemical structure were as shown in Figure 5B. Among them, compound 58 present almost equivalent potency to imatinib in inhibiting STAT1 nuclear translocation. Thus it can be concluded that the newly established HCS assay is able to screening of CSF1R inhibitors in a high throughput way.
Figure 5: Effect of known CSF1R inhibitors on nuclear area, cell area and cell count of CSF1R/GFP-STAT1_U2OS cells. The cells were incubated with varying concentrations of CSF1R inhibitors, GW2580 (A), Sunitinib (B) and Imatinib (C), for 1 hour. Values of normal control group was set as 1. Values are means ± S.D. n=3.
| Compounds | IC50(μM) |
|---|---|
| 34 # | 1.73 ± 0.19 |
| 48 # | 7.64 ± 1.13 |
| 58 # | 0.58 ± 0.01 |
| 63 # | 1.62 ± 0.04 |
Table 1: The IC50 values of the four positive hits in inhibiting GFP-STAT1 nuclear translocation. Values are means ± S.D.
Anti-proliferative activities of the positive compounds on m-nfs-60 cells
The proliferation of murine leukemia cell line M-NFS-60 greatly depends on CSF, and the inhibiting capability of compounds on CSF-1 induced MNFS- 60 cell proliferation was often used in identifying CSF1R inhibitors [11,24]. To further confirm that the results of our screening assay were consistent with traditional screening method, we further examined the anti-proliferative potential of our positive compounds on rhM-CSF induced M-NFS-60 cell proliferation. As shown in Table 2, the growth of M-NFS-60 cells in medium supplemented with 10 ng/mL rhM-CSF was markedly suppressed by treatment with GW2580 (IC50=0.69 ± 0.32 μmol/ L). Similarly, the growth of the cells was also inhibited by Sunitinib, compound 58 and compound 63 with an IC50 of 0.89 ± 0.04 μmol/L, 0.64± 0.03 μmol/L and 1.41 ± 0.2 μmol/L, respectively. However, The IC50 values of Imatinib mesylate, compound 34 and compound 48 for MNFS- 60 cells were all over 3 μmol/L, which exhibited lower activity in the cell proliferation assay.
| Compounds | IC50(μM) |
|---|---|
| GW2580 | 0.69 ± 0.32 |
| Sunitinib | 0.89 ± 0.04 |
| Imatinib mesylate | >3.00 |
| Compound 34 | >3.00 |
| Compound 48 | >3.00 |
| Compound 58 | 0.64 ± 0.03 |
| Compound 63 | 1.41 ± 0.20 |
Table 2: The anti-proliferative activities of CSF1R inhibitor on M-NSF-60 cells. Values are means ± S.D.
Discussion
CSF1R signaling pathway is an important signaling pathway during survival, proliferation and differentiation of macrophages, and its role in the progress of diseases such as cancer, inflammatory diseases as well as Alzheimer’s disease was recognized gradually in recent years. However, targeted drug development remains staged behind. In current paper, a high-content screening assay specialized for screening and identifying of novel CSF1R signaling pathway inhibitors were successfully established. Its accuracy was confirmed by known CSF1R signaling pathway inhibitors, and the efficiency and high throughput were validated by screening of large libraries of available compounds.
The reasonable, reliable and efficient screening method is the key issue in the early stages of drug discovery. Although several methods for detecting potential CSF1R inhibitors had been described previously. However, to some extent, shortcomings at either accuracy or efficiency (i.e. throughput) restricted their reliability and widely utilizing. The most often used time consuming ATP site-dependent competition binding assays involved the expression and isolation of the intracellular domains of human c-Fms kinase from insect cells, and then followed by assaying with ATP and appropriate substrates [26-30]. In cell proliferation assays that also used to detect CSF1R inhibitors, no unified cell types were taken, several kinds of different cell lines including human peripheral blood mononuclear cells (PBMCs), monocytes, macrophages, and M-CSFdependent mouse myeloid M-NFS-60 cells had been used by different institutes [29]. Meanwhile, cellular toxic effect of testing molecules may also affect the accuracy of the assay, which needs independent assay to distinguish. Additionally, as a semi-quantitative method, immunoblotting assays in identifying CSF1R inhibitors by determining the amount of phosphorylated and total CSF1-R after stimulation with M-CSF in human c-Fms kinase stably expressed cells would not be feasible in screening of numerous compounds in a cost-efficient way [17]. Although fluorescent protein and fluorescent dyes such as GFP, EGFP and Hoechst 33342, occasionally were used in evaluating bioactivity of compounds targeting c-Fms kinase in cells but, in general, they were used only in a semiquantitative, low-throughput manner by fluorescence microscopy.
Cellular HCA is an image based quantitative analysis tool developed in recent years, who could measure spatial and temporal transition of interested proteins within cells dynamically or at designated time points . Meanwhile, it is more advantageous than the in-vitro ATP sitedependent competition binding, as multiple cellular parameters based on cell populations can be simultaneously obtained in one test, which provides powerful means in screening of molecular targeted compounds and assessing the quality and efficiency of lead compounds [26]. These advantages of HCA platform have enabled us to develop a HCS assay for rapid screening of compounds that inhibit phosphorylation of human c- Fms kinase. Using a U2OS cell line stably expressing GFP labeled CSF1R downstream protein STAT1, we established a cell line, c-Fms/GFPSTAT1_ U2OS, stably expressing both CSF1R and GFP-STAT1. Based on this, screening of the c-Fms pathway inhibitors can be carried out by simply monitoring the STAT1 translocation from the cytoplasm to the nucleus in cells. This HCS assay uses live cells rather than isolated catalytic domain or full length human of c-Fms kinase and can, therefore, be considered to be more physiologically relevant.
After a routine validation process, we proved that the established cell line exhibit GFP-STAT1 nuclear translocation in response to rhM-CSF stimuli specifically. The accuracy of current estabolished cell model in identifying of c-Fms kinase inhibitor were then confirmed by two marketed CSF1R inhibitors (Sunitinib and Imatinib) and one CSF1R inhibitor (GW2580) under development. As expected, the results showed that the three known CSF1R inhibitors can inhibit GFP-STAT1 nuclear translocation induced by rhM-CSF/c-Fms. The sensitivity of the assay was consistent with other screening assays for pCSF1R and pERK13. And then, the highthroughput of this HCS assay were further validated by using it in screening of a library of 81 newly synthesized CSF1R signaling pathway targeting compounds that designed by chemist in our new drug design center, and 4 compounds were screened out to be effective CSF1R signaling pathway inhibitor. Among them, compound 58 is most potent in inhibiting STAT1 nuclear translocation, with potency almost equivalent to the known CSF1R inhibitor imatinib. The effectiveness were then corroborated with the anti-proliferation assay on the M-NFS-60 cells, a traditional screening method for identifying the CSF1R signaling pathway inhibitors, here compound 58 also showed comparable anti-proliferative bioactivity as sunitinib. Taken together, the newly established CSF1R/GFPSTAT1_ U2OS assay for CSF1R signaling pathway inhibitors was reliable and most importantly, it is easy, rapid, high-throughput, quantitative, and precludes the need for radioactive compounds. However, it's worth noting that "CSF1R inhibitors" may involve M-CSF-binding inhibitors and RTK inhibitors. RTK inhibitors could be detected by current established method, but the detected inhibitors by this method are not always RTK inhibitors. The hits derived from current method also need to be further corroborated by other methods to improve the accuracy. Thus the established HCS assay more suits for preliminary screening of numerous compounds.
In summary, we established a HCS assay for CSF1R signaling pathway inhibitors and validated it in rapid screening of large libraries of available compounds for inhibition of CSF1R signaling pathway. Besides, four hits were identified from 81 compounds and two of them were shown to be able to inhibit the growth of M-NFS-60 cells. Thus the current assay provides a novel approach for CSF1R signaling pathway inhibitors’ screening and identification, and to some extent it will undoubtedly contribute to the progress in finding of new CSF1R signaling pathway inhibitors.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (Grant number 81430090) and the Major Program of the Ministry of Science and Technology of China (Grant numbers 2012ZX09301-001, 2012ZX09301-003). The authors thank Dr. C.J. Sherr and Dr. Martine F. Roussel (Howard Hughes Medical Institute/St. Jude Children’s Research Hospital, Memphis, TN, USA) for their plasmid sincerely.
REFERENCES
- Sherr, Roussel CJ, Rettenmier MF. Colony-stimulating factor-1 receptor (c-fms). Journal of Cellular Biochemistry. 1988;38:179-87.
- Lin H, Lee E, Hestir K, et al. Discovery of a Cytokine and Its Receptor by Functional Screening of the Extracellular Proteome. Science. 2008;320:807-11.
- Stanley ER. The biology and action of colony stimulating factor-1. Stem Cells 1994;1:15-244.
- Varticovski L, Druker B, Morrison D, et al. The colony stimulating factor-1 receptor associates with and activates phosphatidylinositol-3 kinase. Nature. 1989;342:699-702.
- Baccarini M, Sabatini DM, App H, et al. Colony stimulating factor-1 (CSF-1) stimulates temperature dependent phosphorylaation and activation of the RAF-1 proto-oncogene product. Embo Journal. 1990;9:3649-57.
- Yeung YG, Berg KL, Pixley FJ, et al. Protein tyrosine phosphatase-1C is rapidly phosphorylated in tyrosine in macrophages in response to colony stimulating factor-1. Journal of Biological Chemistry. 1992;267:23447-50.
- Novak U, Harpur AG, Paradiso L, et al. Colony-stimulating factor 1-induced STAT1 and STAT3 activation is accompanied by phosphorylation of Tyk2 in macrophages and Tyk2 and JAK1 in fibroblasts. Blood. 1995;86:2948-56.
- Douglass TG, Driggers L, Zhang JG, et al. Macrophage colony stimulating factor: Not just for macrophages anymore! A gateway into complex biologies. International Immunopharmacology. 2008;8:1354-76.
- Hampe A, Shamoon BM, Gobet M, et al. Nucleotide sequence and structural organization of the human FMS proto-oncogene. Oncogene Research. 1989;4:9-17.
- Hume DA, Macdonald KP. Therapeutic applications of macrophage colony-stimulating factor-1 (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood. 2012;119:810-20.
- Mcdonald B, Jansen M, James I, et al. Inhibition of colony-stimulating-factor-1 signaling in vivo with the orally bioavailable cFMS kinase inhibitor GW2580. Proceedings of the National Academy of Sciences. 2005;102:16078-83.
- El Gamal MI, Anbar HS, Yoo KH, et al. FMS Kinase Inhibitors: Current Status and Future Prospects. Medicinal Research Reviews. 2013;33:599-636.
- Uitdehaag JC, Sünnen CM, van Doornmalen AM, et al. Multidimensional Profiling of CSF1R Screening Hits and Inhibitors Assessing Cellular Activity, Target Residence Time, and Selectivity in a Higher Throughput Way. Journal of Biomolecular Screening. 2011;16:1007-17.
- Dewar AL, Zannettino AC, Hughes TP, et al. Inhibition of c-fms by imatinib: expanding the spectrum of treatment. Cell Cycle. 2005;4:851-3.
- Papaetis GS, Syrigos KN. Sunitinib. a multitargeted receptor tyrosine kinase inhibitor in the era of molecular cancer therapies. Biodrugs Clinical Immunotherapeutics Biopharmaceuticals & Gene Therapy. 2009;23:377-89.
- Fabian MA, Biggs WH, Treiber DK, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nature Biotechnology. 2005;23:329-36.
- Guo J, Marcotte PA, Mccall JO, et al. Inhibition of phosphorylation of the colony-stimulating factor-1 receptor (c-Fms) tyrosine kinase in transfected cells by ABT-869 and other tyrosine kinase inhibitors. Molecular Cancer Therapeutics 2006;5:1007-13.
- Leonard WJ. Role of Jak Kinases and STATs in Cytokine Signal Transduction. International Journal of Hematology. 2001;73:271-77.
- Courtneidge SA, Dhand R, Pilat D, et al. Activation of Src family kinases by colony stimulating factor-1, and their association with its receptor. Embo Journal. 1993;12:943-50.
- Ramana CV, Grammatikakis N, Chernov M, et al. Regulation of c‐myc expression by IFN‐γ through Stat1‐dependent and ‐independent pathways. The EMBO Journal. 2000;19:263-272.
- Xiaoxia XU, Min T, Shi W. Improved synthesis of imatinib mesylate. Journal of Pharmaceutical Practice 2014.
- Kai L, Yu-Min DU, Zhao DM, et al. Improved synthesis of sunitinib malate. Chinese Journal of Medicinal Chemistry 2009.
- Emerson HK, Musso DL, Chamberlain SD, et al. Crystal structure of liganded cFMS kinase domain. In US. 2009.
- Nakoinz I, Lee MT, Weaver JF, et al. Differentiation of the IL-3-dependent NFS-60 cell line and adaption to growth in macrophage colony-stimulating factor. Journal of Immunology 1990;145:860-4.
- Zhang JH, Chung TDY, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. Journal of Biomolecular Screening. 1999;4:67-73.
- Giuliano KA, Haskins JR, Taylor DL. Advances in high content screening for drug discovery. Assay & Drug Development Technologies. 2003;1:565-77.
- Carnero A. High throughput screening in drug discovery. Clinical & Translational Oncology. 2006;8:482-90.
- Bickle M. The beautiful cell. high-content screening in drug discovery. Analytical & Bioanalytical Chemistry. 2010;398:219-26.
- Conway BR, Demarest KT. The use of biosensors to study GPCR function: applications for high-content screening. Receptors & Channels. 2002;8:331-41.
- Grånäs C, Lundholt BK, Heydorn A, et al. High content screening for G protein-coupled receptors using cell-based protein translocation assays. Combinatorial Chemistry & High Throughput Screening. 2005;8:301-9.