CYANO RT-Microarray: A Novel Tool to Detect Gene Expression in Cyanobacteria
Received: 23-May-2018 Accepted Date: Jun 26, 2018; Published: 30-Jun-2018
Citation: Medlin LK. CYANO RT-Microarray: A Novel Tool to Detect Gene Expression in Cyanobacteria. J Environ Microbiol 2018;1(1):17-27.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
EU μAQUA made early warning systems for freshwater pathogens and toxins. Barcodes for each cyanobacterial toxin gene published before 2011 were designed and used in a microarray format to capture messenger RNA to detect toxin gene expression at early stages of bloom development. A reverse transcriptase (RT) microarray was developed to detect toxin expression, which had low expression levels. Probes immobilized on the microarray slide captured the mRNA and were extended directly on the microarray. RT extension incorporated fluorescently labeled oligonucleotides to ensure a high signal detected by the microarray scanner. The CYANO RT microarray was laboratory tested with known toxic cyanobacteria and field-tested. Hybridizations without RT extensions were barely detectable in the cultured strains. However, with RT extensions, hybridized mRNA was easily detected. Field samples were equally successful and consistent with companion studies from the same sites using HPLC/(MS-MS) (High Performance Liquid Chromotography/ Mass Spec). In some cases, amplified expression produced a signal with no detection of that toxin using chemical means. The RT microarray may be more sensitive than HPLC/MS-MS. Further studies are needed to determine if the RT-microarray is detecting a very low expression of the toxin genes and, hence, more sensitive as an early warning system predicting the toxin potential.
Introduction
Cyanobacteria are oxygenic phototrophs that produce a variety of toxins, many of which are cyclic peptides; these are hepatotoxins, or neurotoxic alkaloids [1] and pose a serious health threat to drinking water worldwide [2,3]. Hepatotoxins include microcystins, nodularins, and cylindrospermopsins, whereas neurotoxins include anatoxin-A and saxitoxins. Genera typically known to produce these toxins are indicated in Table 1.
Dermotoxins, a third class of toxins, are produced by Lyngbya, Schizothrix, and Oscillatoria [1]. Some cytotoxins are also known [4]. Nutrient availability and other abiotic factors can affect toxin production [5,6]. Different strains of the same species can be toxic or non-toxic, making morphological species identification unreliable for predicting toxins or toxin potential [7]. Functions for the toxins are speculative. They are considered secondary metabolites [1]. Only in a few cases (microcystins and saxitoxins) were the complete pathways known at the time of the design of this microarray [4,8-10]. Anatoxin pathways were published after the toxin array was tested and are not included on this version of the toxin array.
RT-PCR methods have been developed for various toxin genes in various pathways. It has been suggested that these methods could detect cryptic cyanobacterial species, i.e., those capable or potentially producing toxic blooms so that water bodies at potential risk for cyanobacterial toxic blooms could be identified early in the season [11]. However, PCR methods can be biased and subject to inhibition from natural products in the field samples as well as from reaction conditions [12]. If the PCR is inhibited, then positive results can be missed. Our method only uses the RT enzyme once the mRNA has been hybridized to the array (no enzymes used up to this point) and all impurities washed away before the RT enzyme is applied to the microarray.
In keeping with the general objectives of the European Union (EU) μAQUA project to make early warning systems for freshwater pathogens, two microarrays were designed:
• A phylochip to detect pathogenic species in freshwaters, which included cyanobacteria along with other bacteria and protozoa.
• A microarray to detect the messenger RNA (mRNA) from the cyanobacterial toxin genes.
The latter microarray, to detect the expression of the toxin genes, is a new type of microarray and is described in detail here. Probes (barcodes) for each cyanobacterial toxin gene from available publications, prior to 2011, of molecularly characterised cyanotoxin gene pathways (Table 1) were designed and used in a microarray format to capture the mRNA for these genes. The pathway for anatoxin was published after the microarray was designed and tested and is not included here, although these toxins were detected in the two field sites by the chemical methods compared here to the toxin array. The barcodes include 3 coding regions in the aeruginosin pathway, 35 coding regions in the microcystin pathway, 12 coding regions in the cylindrospermopsin cpr/aoa gene cluster pathway, 2 coding regions in the nodularin pathway, 3 coding regions in the saxitoxin pathway, and for controls: 6 coding regions in the phycocyanin pathway, 1 coding region in the gas vesicle pathway, and 7 coding regions involved in housekeeping gene pathways.
| Saxitoxin | Aeruginosin | Microcystin | Cylindropsermopsin | Nodularin | Phycocyanin | Gas Vesicles | Housekeeping Genes |
|---|---|---|---|---|---|---|---|
| Anabaena, Aphanizomenon, Lyngbya, Cylindrospermopsis Al-Tebrineh et al., [22] Fathalli et al., [27] Kellmann et al., [38] |
Planthothrix Microcystis, Nodularia Ballot et al., [23] Fergusson et al., [28] Gugger et al., [30] Halinen et al., [31] Henson et al., [32] Ishida et al., [34] Mihali et al., [42] Rajaniemi et al., [43] |
Microcystis, Anabaena, Planktothrix (Oscillatoria), Nostoc, Dolichospermum Hapalosiphon, Anabaenopsis, Aphanizomenon flos-aquae Briand et al., [25] Fathalli et al., [27] Ginn et al., [29] Hisbergues et al., [33] Jungblut & Neilan [1] Kaebernick et al., Noguchi et al., [47] Nonneman & Zimba [48] Ostermaier & Kurmayer Ouahid et al., [52] Rantala et al., [54] Sipari et al., [63] Tillett et al., [63] Tooming-Klunderud et al., [64] Vaitomaa et al., [65] Valerio et al. |
Cylindrospermopsis, Aphanizomenon, Anabaena Raphidiopsis, Umezakia Ballot et al., [23] Rasmussen et al., [56] Schembri et al., [59] Valério et al., [66] Wilson et al., [68] |
Nodularia Jonasson et al., [36] Jungblut & Neilan [37] Koskenniemi et al., [39] Lyra et al., [41] Moffitt & Neilan [43] Moffitt et al., [43] |
Ballot et al., [23] | Becker et al., [24] | Iteman et al., [35] Neilan et al., [45] Nübel et al., [49] Rudi et al., [57] Schoenhuber et al., [60] Svenning et al., [62] |
Table 1: Summary of the literature [23-68] used to assemble and modify the probes designed for the toxin array. Pathways published after 2011 were not included in this version of the microarray. The cyanobacterial genera typically associated with each toxin are indicated.
Initially, the signals obtained from the hybridizations were so low that they could barely be distinguished above the background signal and so a method was developed to extend the probes (barcodes) using reverse transcriptase directly on the microarray by incorporating fluorescently labeled oligonucleotides as it was extended. This is essentially the same reaction that is performed to generate complementary DNA (cDNA) only it was performed directly on the microarray to produce an enhanced signal that enabled the mRNA to be detected from a variety of cyanotoxin genes. This novel tool is a measure of potential toxigenicity in cyanobacteria and could possibly be used to predict toxic cyanobacterial blooms or to identify the potential of any water body for cyanobacterial blooms, especially if used routinely as a monitoring tool and used in parallel with a species array [13-16] to compare the species present with their potential toxicity. Initial tests are presented here to show its feasibility and field tests from one sampling day from the Netherlands are reviewed [16] and compared to field samples taken for two years in France.
Material and Methods
Environmental sites
The toxin gene array was laboratory tested using pure cultures of cyanobacterial species known to be toxic or nontoxic and field-tested in two countries (the Netherlands and France, Table 2). In these two countries, aliquots of the same sample were tested for toxins using standard methods in companion papers [16-18]. In the Netherlands, six water bodies were sampled once in the summer of 2015, and in France, Canet Lagoon (Table 2) was sampled monthly from August 2011 to May 2013. The Dutch sites LA1–LA3 are along an inland waterway and sites LA4–LA6 are small lakes; all have known cyanobacterial blooms each year, primarily from Dolichospermum, Microcystis, and Aphanizomenon. The Anabaena probes recognise Dolichospermum, which was formerly placed in Anabaena. The Canet site was selected in the μAQUA project as a representative brackish water site in this region of France, and its cyanobacterial community was unknown at the time of sampling.
| Code | Location | Latitude | Longitude |
|---|---|---|---|
| LA1 | Nuldernauw Harbor | 52°26′ | 5°47′ |
| LA2 | Nuldernauw | 52°26′ | 5°49′ |
| LA3 | Wolderwijd Zoetermeerse | 52°33′ | 5°55′ |
| LA4 | Plas | 52°08′ | 4°51′ |
| LA5 | De Put De Grote | 52°08′ | 4°51′ |
| LA6 | Plas | 52°05′ | 4°32′ |
| CA | Canet Lagoon, | 42°66′ | 3°03′ |
Table 2: Location of the sampling sites from the six Netherlands lakes (LA) and Canet Lagoon, France (CA).
Filtration for RNA extraction
A total of 50 L was collected from the environmental sites sampled, concentrated using the Hemoflow hollow fiber filter (Hemoflow HF80S, Fresenius, Bad Homberg, Germany), and back-flashed with 1 L solution (1 L milliQ-H2O, phosphate buffered saline (0.01 M), 5 mL Tween 80 solution (0.5%), 100 mg Sodium Hexametaphosphate (0.01%), 1 mL Antifoam B emulsion) to yield a 1 L concentrate. Two hundred milliliters of the concentrated eluate were sequentially filtered through the eight different filters of decreasing pore size (20 μm, 10 μm, 5 μm, 2 μm, 0.8 μm, 0.45 μm, 0.1 μm, and 0.025 μm filters, Millipore, Billerica, MA, USA) and placed into 1 mL of Tri-Reagent (Sigma Aldrich, St. Louis, MO, USA) [13], then stored frozen at −80 °C for further analysis and extraction for downstream analysis by the species and the toxin array. The hollow fibre filters are very easy to use, making the filtration of 50 liters attainable in 30 minutes or less. This is an obvious advantage over the standard filtration of one liter or less using standard Millipore filtration methods. Such a large volume is needed for the extraction of total RNA for the species microarray and for sufficient mRNA typical of low expression of toxic genes. For small water bodies or rivers a 50 liter sample should be more than adequate to characterize the water body but for larger water bodies multiples of the 50 liter samples could be easily taken and filtered in the same day.
RNA extractions
All filters were pooled and extracted with Tri-Reagent following the protocol in Lewis [19] as modified in Kegel [20] with the following additional modifications for cyanobacterial extractions from Yilmaz [21]. Briefly, after addition of the RNA extraction control (500,000 Dunaliella cells) and 250 μL of 0.5 mm ZR BashingBeads™ to the pooled samples, samples were bead-beaten twice at maximum speed for 1 min. Then, 0.2 M Tris (pH 5.0), 0.02 M ethylenediaminetetraacetic acid (EDTA) (pH 8.0), and 1% sodium dodecyl sulfate (SDS) buffer were added and well mixed followed by the addition of 0.75 M NH4 acetate and 1% potassium ethyl xanthogenate (Sigma, St. Louis, MO, USA) and incubation at 65°C for 15 min. The following procedure was then performed as described in Kegel [20].
Probe design
A microarray for pathogenic freshwater species was developed and has been tested elsewhere within the EU μAQUA project [14-17,22]. A companion microarray for toxin pathway genes was also developed and this method and preliminary field tests of this array are presented here. For the toxin array, probes were designed to target the microcystin synthetase gene cluster, including ten genes (mcyA to J), aeruginosin synthetase gene clusters (aer), saxitoxin biosynthesis gene cluster (sxt), and the aoa/cyr clusters putatively involved in cylindrospermopsin biosynthesis (Table 2). These probes were spotted onto the toxin array to capture the messenger rRNA (mRNA) for these toxin genes. Toxin pathways published after 2011 were not included. Probes were tested for specificity using various cyanobacterial species including Cylindrospermopsis raciborski, Aphanizomenon PCC 7905, Microcystis sp. BC 84/1, Microcystis aeruginosa CCAP 1450/8, Planktothrix agardhii strains 34, 126, and 137, Planktothrix rubescens strains 34, 137, and 9316, and Dolichospermum sp. (formerly Anabaena sp.). All probe sequences are patent pending.
Microarray hybridization for the gene expression array
The microarrays used for the toxin gene expression detection were spotted with 83 probes, including 3 positive controls for hybridization quality, 3 negative controls for absence of non-specific hybridization, 7 positive controls for RNA extraction efficiency, and 70 probes encompassing different coding regions of genes involved in toxin biosynthesis and housekeeping gene pathways as follows: 3 coding regions in the aeruginosin pathway, 35 coding regions in the microcystin pathway, 12 coding regions involved in the cylindrospermopsin toxin pathway, 2 coding regions involved in the nodularin pathway, 6 coding regions involved in the phycocyanin pathway, 1 coding region involved in the gas vesicle pathway, 3 coding regions involved in the saxitoxin pathway, and 7 coding regions involved in housekeeping genes pathways. The non-toxin genes were added to the microarray as controls for cyanobacterial populations. These probes were modified for microarray use from the references [23-68] in Table 1 according to each toxin pathway.
Hybridization of the mrna to the toxin array
Each microarray slide contained two arrays with eight replicates for each probe. Hybridizations of each sample were performed on different slides, thus producing a pseudo-replicate. Considering two arrays per sample, each probe is, therefore, represented by 16 spots, and the signal for the 16 spots was averaged. One mL of the mixture obtained from the pooled filters previously stored in TRI-reagent at −80°C degree plus an internal extraction quality control (Lambda DNA) was processed for total RNA extraction using TRlzol® Reagent according to the patented MIDTAL procedure (patent WO2015008011 A1). RNA quality and purity (260/280 ratio: 1.8–2.2 and 260/230 ratio: 1.8–2.3) were measured by NanoDrop® Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The integrity and size distribution of total RNA was checked with a 2100 Bioanalyzer (Agilent, Technologies Inc. Santa Clara, CA, USA). One microgram of total RNA extracted from field samples was labeled and purified using a Platinum Bright 647 Infrared Nucleic Acid kit (Leica Biosystem, Nussloch GmbH, Nußloch, Germany) according to the manufacturer’s instructions. The degree of labeling (DoL) was determined by measuring concentration and incorporating the dye using a NanoDrop® Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Samples with DoL values between 1.0 and3.0 were processed to hybridization. Labeled RNAs were fragmented by adding 1/10 volume of Fragmentation Buffer (salt buffer) (100 mM ZnCl2 in 100 mM Tris-HCl, pH 7.0) and incubated for 15 min at 70°C and immediately chilled down on ice to room temperature. The reaction was stopped by adding 1/10 volume of 0.5 M EDTA, pH=8 to the sample. Microarray hybridizations were performed following optimized procedures based on protocols published in Kegel et al. [13]. Briefly, labeled field samples (1 μg RNA) were mixed with (2 ×) hybridization buffer containing 3 μL Poly-dA (1 μM) and 10 ng TBPcontrol made up to a final volume of 30 μL. Poly-dA was added to block the poly-T spacer on the probe and TBP was the TATA box gene fragment added as the positive hybridization control [15]. The labeled RNA was then denatured for 5 min at 95°C. After denaturation, the samples were placed on ice, and 7.5 μL of 4X KREA block (background blocker from Leica Biosystems, Nussloch GmbH, Nußloch, Germany) were added. The hybridization mixture was equally distributed to each array covered with lifter-slips (cover slips with raised edges) and cleaned with ethanol (LifterSlips, Erie Scientific, VWR International, Radnor, PA, USA). Slides were placed into SCIENION’s sciHYBCHAMBER that maintained a moist environment to avoid evaporation, and hybridizations were carried out for 1 h at 65°C using a water bath. To remove unhybridised RNA, the slides were successively washed with three washing steps with increasing buffer stringency under agitation and in the dark to protect the fluorophore. The first buffer (2 × saline-sodium citrate (SSC)/10 mM EDTA/0.05% SDS) and the second buffer (0.5 × SSC/10 mM EDTA) washings were performed at room temperature for 10 min, whereas the final most stringent wash (0.2 × SSC/10 mM EDTA) was performed at 50°C.
The signals from the two replicate hybridization arrays are regressed one against the other in order to obtain the Cy5 RNA hybridization and signal amplification efficiency. The closer R2 is to 1, the more efficient and reliable the hybridization was between the two blocks. Values under 75% were repeated, if necessary (Table 3).
| Sample date | R2 |
|---|---|
| 05-07-2011 | ND |
| 09-08-2011 | ND |
| 26-09-2011 | 0.98 |
| 24-10-2011 | 0.94 |
| 07-03-2012 | 0.99 |
| 03-04-2012 | 0.92 |
| 09-05-2012 | 0.97 |
| 08-06-2012 | 0.99 |
| 03-07-2012 | 0.94 |
| 07-08-2012 | 0.98 |
| 07-08-2012 | ND |
| 18-09-2012 | 0.99 |
| 16-10-2012 | 0.93 |
| 19-11-2012 | 0.96 |
| 28-03-2013 | 0.97 |
| 18-04-2013 | 0.99 |
| 16-05-2013 | 0.9 |
Table 3: R2 of the correlation/comparison of replicate hybridizations of Canet-Saint Nazaire samples for each sampling date over the two year period. Examples of the replicate hybridization regression curves are shown in Figure 1.
RT extension of mRNA bound to microarray
After the third wash of the microarray, lifter-slips were put back onto each array and retro-transcription was performed using the SuperScript® VILO™ cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA). Three washings of the microarray should ensure that any potential inhibitors of the RT enzyme would be removed. The reaction was performed following the manufacturer’s instructions with the addition of Cy5dCTP to the master mix in a final volume of 30 μL, and the mixture was incubated for 45 min at 50°C. The slides were washed successively with the washing buffers 1, 2, and 3 as described above.
Microarray analysis
GPR Files exported from the Genepix scanner were loaded into the GPR analyzer program (69) for analysis of the replicate hybridizations. Results were exported from the GPR analyzer program into Excel and normalized with the signal from the buffer. Some Excel files were imported into PermutMatrix, which is freely available software that allows a heat map representation of microarray data (http://www.atgc-montpellier.fr/permutmatrix/).
UPLC/MS-MS/microspheres
All of the chemical analyses shown in this paper are detailed in Greer et al. [17,18]. The results in those papers and those have been reproduced here for comparison with the toxin array from Canet Lagoon [16].
Results
Hybridization quality and experimetnal replication
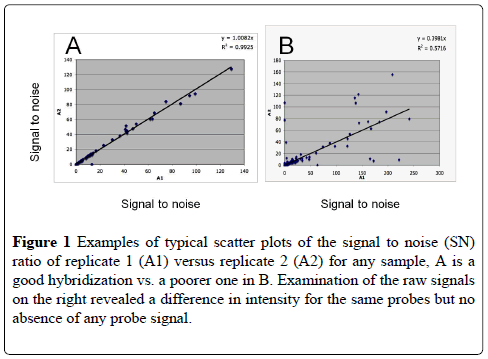
The hybridization and signal amplification quality of each sample was checked by regression scatter plotting the SN (signal to noise) ratio of each hybridization replicate. The average curve and the R2 were calculated for each sample and any sample showing an R2 of less than 0.75 was reexamined to see why the correlations were not over .90 was discarded and redone. Examples of a good and a poorer hybridization scatter plots are shown in Figure 1. Of the nineteen samples taken over the two year period, only four were not processed for analysis because of poor RNA quality. All R2 values were over 0.9 (Table 3).
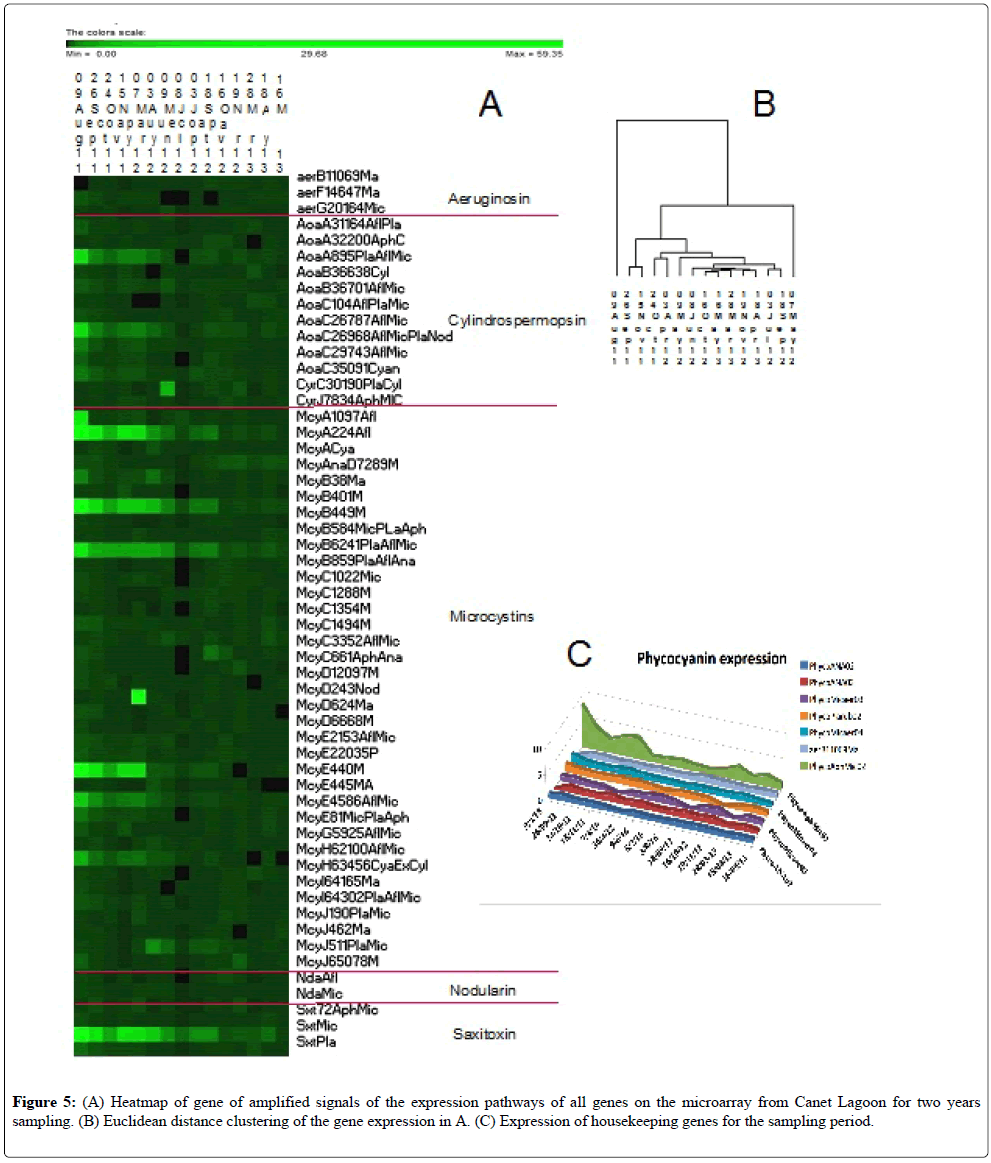
Figure 1: Examples of typical scatter plots of the signal to noise (SN) ratio of replicate 1 (A1) versus replicate 2 (A2) for any sample, A is a good hybridization vs. a poorer one in B. Examination of the raw signals on the right revealed a difference in intensity for the same probes but no absence of any probe signal.
Laboratory tests
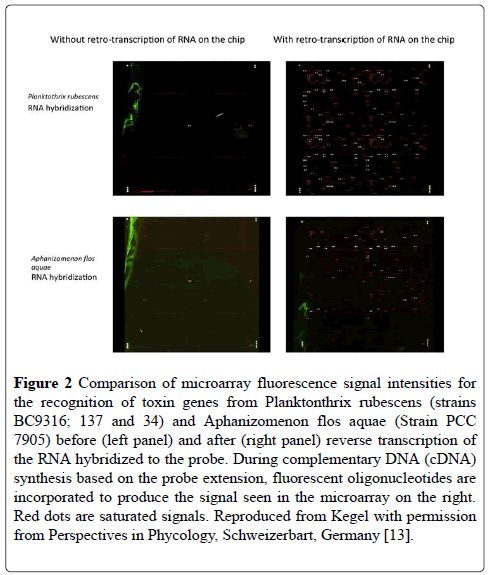
Figure 2 shows a comparison of before and after reverse transcriptase (RT) amplification of the captured mRNA for the toxin genes isolated from two cultures of different cyanobacterial species. It is immediately obvious that the RT amplification has greatly increased the signal intensity to a level at which it could be detected by the microarray scanner (Figure 2).
Figure 2: Comparison of microarray fluorescence signal intensities for the recognition of toxin genes from Planktonthrix rubescens (strains BC9316; 137 and 34) and Aphanizomenon flos aquae (Strain PCC 7905) before (left panel) and after (right panel) reverse transcription of the RNA hybridized to the probe. During complementary DNA (cDNA) synthesis based on the probe extension, fluorescent oligonucleotides are incorporated to produce the signal seen in the microarray on the right. Red dots are saturated signals. Reproduced from Kegel with permission from Perspectives in Phycology, Schweizerbart, Germany [13].
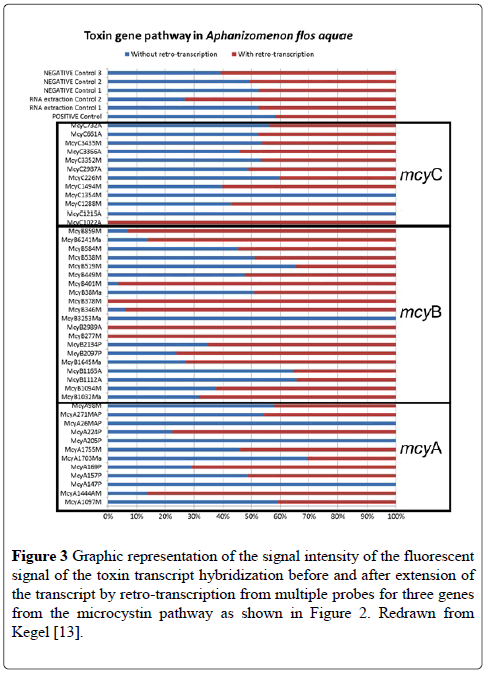
Among the pathways studied, several were housekeeping genes specific or typical of cyanobacteria, which were added to the microarray as positive controls. These included genes for gas vesicle proteins and for phycocynanin. Although these control probes are highlighted between the two species in Figure 2, it is clear that the mRNA captured by the microarray is different between the two species because of the different toxins that are produced by the two species tested. Figure 3 shows the % difference in the signal intensity for the genes on the microarray for mcyA, mcyB and mcyC from A. flosaquae before and after reverse transcription directly on the microarray (Figure 3).
Figure 3: Graphic representation of the signal intensity of the fluorescent signal of the toxin transcript hybridization before and after extension of the transcript by retro-transcription from multiple probes for three genes from the microcystin pathway as shown in Figure 2. Redrawn from Kegel [13].
Field tests
The netherlands lakes
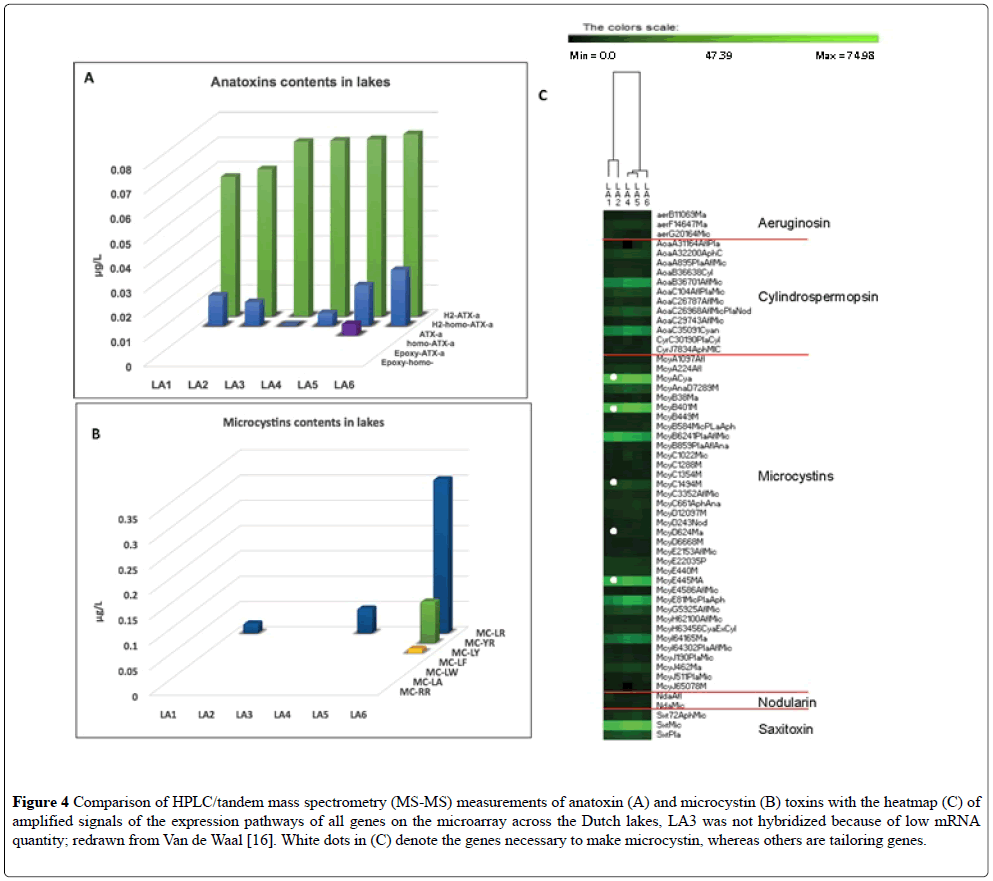
Six shallow water bodies were sampled once during the summer of 2013 (Table 4, Figure 1) [16]. The extraction of RNA from the sample taken at LA 3 did not yield sufficiently high quality RNA for both the species array and the toxin array to be performed, so only the hybridization for the species array was performed (unpublished). In the Van der Waal study [16], toxins were also measured by a standard method (UPLC) and only Anatoxin-A and some microcystins were above the detection level (Figures 4A and 4B). In all of lakes tested with the toxin array, expression of the toxin genes was recorded (Figure 4C) but not for all toxin pathways. The toxin profiles in the interconnected waterways LA1-LA2 were more similar than to that of the isolated lakes. The mcyA, B, E, G, and J genes, and the AoaB, AoaC, and saxitoxin genes were actively expressed as compared to other genes in the other toxin pathways on the microarray. Saxitoxin was not detected by the traditional methods of UPLC [17], but its mRNA was detected in the toxin array. The highlighted probes mainly concerned Aphanizomenon, Anabaena (represented in the Dutch water bodies as Dolichospermum), and Microcystis toxin genes and were present in all sites but with relatively higher signals in site LA4 and LA5 [17]. In the microcystin pathway, mcyC and mcyD were barely detectable in the gene expression array, but mcyA and mcyE displayed very high signals, Figure 4C. Genes mcyA–E are required to produce the toxins, whereas mcyG–J are involved in toxin modification, i.e., tailoring genes. Thus, there was a good correlation between toxins recorded either by the toxin array or by traditional methods and the species array [17].
| Date | Anatoxin | Aeruginosin | Microcystin | Microcystin | Cylindrospermopsin | Cylindrospermopsin | Nodularin | Saxitoxin | Phycocyanin | Housekeeping |
|---|---|---|---|---|---|---|---|---|---|---|
| A-E genes | H,I J genes | cyrA–O genes | aoaA–C genes | |||||||
| 09-08-2011 | nt | RT | RT | RT | RT | RT | RT | RT | RT | RT |
| 26-09-2011 | nt | RT | RT | RT | RT | RT | RT | RT | RT | RT |
| 24-10-2011 | M, H | RT | RT | RT | RT | RT | RT | RT | RT | RT |
| 15-11-2011 | M | RT | RT | RT | RT | RT | RT | RT | RT | RT |
| 07-03-2012 | M | RT | RT | RT | RT | RT | RT | RT | RT | RT |
| 03-04-2012 | M, H | RT | RT | RT | RT | RT | RT | RT | RT | RT |
| 09-05-2012 | M | RT | RT | RT | RT | RT | RT | RT | RT | RT |
| 08-06-2012 | M, H | RT | RT | RT | RT | RT | RT | RT | RT | RT |
| 03-07-2012 | M | RT | RT | RT | RT | RT | RT | RT | RT | RT |
| 18-09-2012 | M | RT | RT | RT | RT | RT | RT | RT | RT | RT |
| 16-10-2012 | M | RT | RT | RT | RT | RT | RT | RT | RT | RT |
| 19-11-2012 | M | RT | RT | RT | RT | RT | RT | RT | RT | RT |
| 28-03-2013 | M, H | RT | RT | RT | H, RT | H, RT | RT | RT | RT | RT |
| 18-04-2013 | M, H | RT | RT | RT | H, RT | H, RT | RT | RT | RT | RT |
| 16-05-2013 | M, H | RT | RT | RT | RT | RT | RT | RT | RT | RT |
Table 4: Comparison of microsphere data (M) [17] and UPLC-MS/MS data (H) [17,18] with reverse transcriptase (RT) gene expression from Canet Lagoon. Gene expression values over 1.5 are in bold. nt=not tested. For the two cylindrospermopsin pathways, the chemical detection is reported in both pathways although it cannot distinguish the two pathways.
Figure 4: Comparison of HPLC/tandem mass spectrometry (MS-MS) measurements of anatoxin (A) and microcystin (B) toxins with the heatmap (C) of amplified signals of the expression pathways of all genes on the microarray across the Dutch lakes, LA3 was not hybridized because of low mRNA quantity; redrawn from Van de Waal [16]. White dots in (C) denote the genes necessary to make microcystin, whereas others are tailoring genes.
Canet saint nazaire
This coastal lagoon exhibits a wide variation in salinity (9 to 28 PSU average range according to Casabianca (70). No obvious cyanobacterial blooms were observed at the Canet Saint Nazaire site during the sampling period.
Strong signals were obtained for some toxin pathways, emphasizing the toxic potential of the cyanobacteria sampled throughout the sampling period, being stronger in 2011 than in 2012-13 (Figure 5). The level of housekeeping gene expression was also moderate to low, no more than 10 times above the background level, which likely indicates relatively inactive cells (Figure 5C) or a low number of cells. This point emphasizes the need to use this tool as a monitoring device to capture and correctly interpret the population status of the cyanobacterial community, monitoring them initially as low numbers (low signal) to high numbers with increasing expression of toxin genes (high signal) to bloom decline and many senescent cells (low signal). Continual monitoring would enable the correct interpretation of the signal.
Nodularin is a toxin produced by Nodularia using the NdaA–I gene cluster. The nodularin pathway was found to be continuously slightly active during all of the sampling campaign showing no peaks in abundance (Figure 5, Table 4). Nodularia spumigena, the causative species producing this toxin, was identified by the ribosomal RNA (rRNA) species microarray but also isolated into culture in the spring and autumn periods (unpublished).
Aeruginosin can be produced by Nodularia, Microcystis and Plantothrix. The three genes on the toxin array were expressed in similar low quantities throughout the sampling period (Figure 5, Table 4) and could have been produced by Nodularia because of its isolation during the sampling period [18].
Cylindrospermospin is produced by numerous cyanobacteria including Cylindrospermospis, Anabaena, Oscillatoria, Raphidiopsis curvata, Umezakia natans, and Aphanizomenon. This toxin is produced by two gene clusters: cyrA–O and/or aoaA–C. The cylindrospermopsin toxin pathway did not show any cyr gene expression except for one sample collected on 9 August 2011 (Table 4). However, the Aoa gene cluster showed a high expression level for the first period of the sample collection spanning from August 2011 to August 2012 (Figure 5, Table 3). UPLC/MS-MS found small traces of the toxin in two samples in the Spring of 2013.
Saxitoxin is encoded by the sxtA–Z genes and is found in Anabaena circinalis, Aphanizomenon flosaqua, Aphanizomenon grazile, Cylindrospermopsis raciborskii, and Lyngbya wollei. Expression of genes coding for this neurotoxin was found in the Canet Saint Nazaire samples with particularly high signals in the first sampling year (Figure 5, Table 4). This neurotoxin was not measured in accompanying UPLC/MS-MS detection studies [18].
Anatoxins were consistently detected by the microsphere or Luminex method, but less so by Ultra-Performance Liquid Chromatography (UPLC)- tandem mass spectrometry (MS-MS) [17,18]. On three occasions, they were above 5 mg/L, but this pathway is not on the present version of the microarray (Table 2). These toxins were detected in Oct of 2011 and in the spring of 2012 and 2013.
Genes for microcystin synthesis and modification were highly expressed especially in the first sampling year. No microcystin toxins were detected by chemical means [17,18]. The highest gene expression of the microcystin biosynthesis pathway involving the McyA–J gene cluster were the microcystin A, B and E genes, but the other microcystin genes required to produce the toxins were present only at expression level throughout the sampling period, supporting the potential toxicity of Microcystis, Anabaena, and Aphanizomenon (Figure 5, Table 4) who were found by the species array (Medlin unpublished).
These gene expression patterns tended to support the toxin detection by chemical means in that in no case were the toxins confirmed by the chemical detection of the toxins themselves by UPLC/MS-MS nor by the microsphere methods [17,18] that were not present on the toxin array. This excludes the detection of anatoxins, whose pathway was published after the toxin array was designed and spotted. In the Canet samples, the chemical analyses using UPLC-MS/MS [17,18] detected small amounts of H2-ATXa and some traces of H2-homo-ATX-a in 3 samples, whereas significant levels of both were detected in all samples using the more sensitive microsphere or Luminex technology. Microcystins were not detected in seawater and brackish water samples as compared to freshwater samples and it was concluded that the UPLC/MS-MS detection suffered from a matrix effect in seawater and brackish water samples (i.e., interference from solutes in the field sample that cannot be removed during the extraction process) as compared to Luminex results [17,18].
A Euclidean distance tree was constructed from the signal intensities (Figure 5B). The two outliers correspond to the two dates with the strongest mycrocystin signals. After that the sampling dates tended to cluster by seasons or sequential dates, with the samples from 2011 in two clusters followed by the 2012-2013 samples.
Discussion
The presence of cyanobacteria in aquatic environments leads to the potential risk of cyanotoxin contamination of the environment and to public health. The early detection of cellular growth and bloom monitoring can be measured by cell counts or other means of molecular detection. Measurements of cell densities or pigment contents allow both, but these methods are not sufficiently accurate to predict actual cyanobacterial risk. Even detection by PCR or qPCR may not be a reliable indicator of cyanobacterial toxin risk [71]. Our novel microarray allows for the detection of very low expression of mRNA from cyanobacterial toxin genes by amplifying that signal using RT directly on the microarray. This is the first attempt to use RT directly in a microarray format. A reverse transcriptase reaction can be performed directly on the microarray to extend the mRNAs bound to the capture barcodes on the microarray so that they produce a signal that can be read by the laser. The variation in signal intensity would suggest that the signal intensity is at least semi-quantitative, but this remains to be fully tested. This effectively extends the use of a microarray to detect genes that are expressed in low quantities and provides a new tool for early warning of the toxic potential of any water body. Preliminary field tests suggest that it is more sensitive than the chemical tests, because, in the field tests, there were samples in which cells were detected by microscopic counts (16 Van der Waal et al. 2018) where the toxin array detected gene expression of the toxins, but there was no detection by chemical means. This method has an advantage over qPCR methods in that all genes can be detected simultaneously, and the isolation of the mRNA prior to the RT amplification of the signal does not involve any enzymes. qPCR would only detect the presence of toxin genes and not if they were being actively expressed. Natural PCR inhibitors are known to affect positive results [69-72]. Cost comparisons to HPLC are unknown.
For the Netherlands samples taken on a single day in the summer of 2015, the toxin array results were consistent with the species microarray and cell count results, which showed the presence of the corresponding toxin producer [16] and represent only a snapshot in time. The toxin array detected the presence of all of the toxins detected by UPLC/MS-MS [17,18], except for the anatoxins,, which are not on the toxin array. In Microcystis, cell counts were high in some lakes where the toxin array showed low expression and in Aphanizomenon, gene expression was high where no cells were seen. In that study, low viability was used to explain the first discrepancy, whereas greater sample volumes (20L) as compared to 1 mL were used to explain the second anomaly between cell counts and gene expression.
Notably in the gene cluster for microcystins, McyC and McyD were expressed in lower amounts, and their microarray signal was just above the detection level in all of the Netherlands lakes. This would suggest either that the population had finished producing its toxins and was becoming senescent, or that these gene products are needed in a lower concentration than the other components of the gene cluster to make the active toxin; both of these explanations are supported by studies on the genetic basis of toxin production [7]. In contrast, Antoxins must have been highly expressed because high amounts of toxins were detected by UPCA/MS-MS, but cell numbers inferred from the microarray species array were notably lower than the actual cell numbers counted [16]. One interpretation of this could be that there was a subset of the Anabaena population that was actively growing and producing the toxins, or that the population was dying and that toxin production increased as the cells became senescent.
Gene expression for one gene in the saxitoxin cluster was detected, but no cells capable of producing that toxin were detected, nor were the toxin itself detected chemically. More genes would be needed on the array for each of the genes present for this toxin.
For the Canet Lagoon, no cell counts were made, and only a comparison between chemical analyses and microarray analysis is possible. Several genes in the cylindrospermopsin pathway were detected by the array, but no toxins were detected with either the microspheres or trace amounts were detected with the UPLC methods [17]. Nodularin gene expression was consistent also with the detection of the causative species and its detection on the species array (unpublished), but the toxin was not detected in the two samples measured by UPLC in the companion study made by Greer et al. [17]. Low expression of the genes was detected by the microarray. Relatively high expression of saxitoxin was not confirmed by UPLC/MSMS tests [17]. These observations would suggest that the toxin array is more sensitive and can detect toxin potential before it can be detected chemically.
The toxin chip is only qualitative but reveals the expression of genes involved in each toxin biosynthesis pathway tested and, in some cases, revealed them before they were detected by chemical means. The toxin chip can serve as a potential early warning system in any monitoring study, which qPCR could only show that species with toxin genes are present. It is clear from these preliminary studies that the toxin array shows great potential. A more in-depth study should be made, making a direct comparison of the gene expression array with toxin measurements by standard methods (UPLC, etc.) with laboratory cultures so that limits of detection and quantification (LOD; LOQ) can be determined. Also, a time series study should be performed in any water body to determine how much in advance of the chemical detection the microarray can detect gene expression of the toxins. The toxin array should be studied in more detail with more toxic cultures and concomitant chemical analysis to determine if the array can detect the expression of the genes before they can be detected chemically. Should this prove to be the case, then the toxin array is more sensitive and could provide an earlier detection of the toxic potential of any water body in any monitoring scheme.
Acknowledgements
This research was funded by EU FP 7 μAQUA contract No. 265409.
REFERENCES
- Kaebernick M, Neilan BA, Borner T, et al. Light and the transcriptional response of the microcystin biosynthesis gene cluster. Appl Environ Microbiol. 2000;66:3387-92.
- Stewart I, Webb PM, Schluter PJ, et al. Recreational and occupational field exposure to freshwater cyanobacteria-A review of anecdotal and case reports epidemiological studies and the challenges for epidemiologic assessment. Environ Health. 2006;5:6.
- Wood R. Acute animal and human poisonings from cyanotoxin exposure-A review of the literature. Environ Int. 2016;91:276-82.
- Pearson L, Mihali T, Moffitt M, et al. On the chemistry toxicology and genetics of the Cyanobacterial toxins: Microcystin, Nodularin, Saxitoxin, and Cylindrospermopsin. Mar Drugs. 2010;8:1650-80.
- Sivonen K, Jones G. Cyanobacterial toxins. In: Chorus I, Bartram J (editors). Toxic Cyanobacteria in Water. A Guide to Their Public Health Consequences Monitoring and Management, London: E and FN Spoon, 1999,41-111.
- O'Neil JM, Davis TW, Burford MA, et al. The rise of harmful cyanobacteria blooms: The potential roles of eutrophication and climate change. Harm Algae. 2012;14:313-34.
- Kuramyer R, Chrsitensen G, Fastner J, et al. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ Microbiol. 2004;6:831-41.
- Dittmann E, Neilan BA, Erhard M, et al. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol Microbiol. 1997;26:779-87.
- Marahiel MA. Multidomain enzymes involved in peptide synthesis. FEBS Lett. 1992;307:40-3.
- Nishizawa T, Asayama M, Fujii K, et al. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J Biochem. 1999;126:520-26.
- Kurmeyer R, Christiansen G. The genetic basis of toxin production in cyanobacteria. Freshw Rev. 2009;2:31-50.
- Miranda JA, Stewart GF. Variables influencing the efficiency and interpretation of reverse transcription quantitative PCR (RT-qPCR) an empirical study using Bacteriophage MS2. J Virol Methods. 2017;241:1-10.
- Kegel JU, Guillebault D, Medlin LK. Application of microarrays (phylochips) for analysis of community diversity by species identification. Perspect Phycol. 2016;9:93-106.
- Baudart J, Guillebault D, Mielke E, et al. Microarray (phylochip) analysis of freshwater pathogens at several sites along the Northern German coast transecting both estuarine and freshwaters. Appl Microbiol Biotechnol. 2016;101:871-86.
- Akcaalan R, Albay M, Koker L, et al. Seasonal dynamics of freshwater pathogens as measured by microarray at Lake Sapanca a drinking water source in the north-eastern part of Turkey. Environ Monit Assess. 2017;212.
- Van der Waal D, Guillebault D, Alfonso A, et al. Aqua microarrays for phylogenetic and toxin expression of cyanobacteria with validation by cell counts and UPLC/MS-MS. Harm Algae 2018.
- Greer B, McNamee SE, Boots B, et al. A validated UPLC-MS/MS method for the surveillance of ten aquatic biotoxins in European brackish and freshwater systems. Harmful Algae. 2016;55:31-40.
- Rodriguez I, Alfonso C, Alfonso A, et al. Toxin profile in samples collected in fresh and brackish water in Germany. Toxicon. 2016;91:35-44.
- Kegel JU, Medlin LK. Introduction to project MIDTAL its methods and samples from Arcachon Bay France. Environ Sci Pollut Res. 2013;20:6690-6704.
- Yilmaz M, Philips E, Tillett D. Improved methods for the isolation of cyanobacterial DNA from environmental samples. J Phycol. 2009;45:517-21.
- Medlin LK, Guillebault D, Mengs G, et al. New molecular tools: application of the AQUA phylochip and concomitant FISH probes to study freshwater pathogens from samples taken along the Tiber River Italy. In: River Basin Management IX, Brebbia CA, Boukalova Z (editors), WIT Trans Ecol Environ. 2017;221:109-22.
- Al-Tebrineh J, Mihali TK, Pomati F, et al. Detection of saxitoxin-producing cyanobacteria and Anabaena circinalis in environmental water blooms by quantitative PCR. Appl Environ Microbiol. 2010;76:7836-42.
- Ballot A, Fastner J, Wiedner C. Paralytic shellfish poisoning toxin-producing cyanobacterium Aphanizomenon gracile in Northeast Germany. Appl Environ Microbiol. 2010;76:1173-80.
- Becker S, Hayes PK, Walsby AE. Different gvpC length variants are transcribed within single filaments of the cyanobacterium Planktothrix rubescens. Microbiol. 2005;151:59-67.
- Briand E, Escoffier N, Straub C, et al. Spatiotemporal changes in the genetic diversity of a bloom-forming Microcystis aeruginosa (cyanobacteria) population. ISME J 2009;3:419-29.
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem1987;162:156-59.
- Fathalli A, Jenhani ABR, Moreira C, et al. Molecular and phylogenetic characterization of potentially toxic cyanobacteria in Tunisian freshwaters. Syst Appl Microbiol. 2011;34:303-10.
- Fergusson KM, Saint CP. Molecular phylogeny of Anabaena circinalis and its identification in environmental samples by PCR. Appl Environ Microbiol. 2000;66:4145-48.
- Ginn HP, Pearson LA, Neilan BA. NtcA from Microcystis aeruginosa PCC 7806 is autoregulatory and binds to the microcystin promoter. Appl Environ Microbiol. 2010;76:4362-68.
- Gugger M, Lyra C, Henriksen P, et al. Phylogenetic comparison of the cyanobacterial genera Anabaena and Aphanizomenon. Int J Syst Evol Microbiol. 2002;52:1867-80.
- Halinen K, Fewer DP, Sihvonen LM, et al. Genetic diversity in strains of the genus Anabaena isolated from planktonic and benthic habitats of the Gulf of Finland (Baltic Sea). FEMS Microbiol Eco. 2008;64:199-208.
- Henson BJ, Watson LE, Barnum SR. Molecular differentiation of the heterocystous Cyanobacteria (Nostoc and Anabaena) based on complete Nif D sequences. Curr Microbiol. 2002;45:161-64.
- Hisbergues M, Christiansen G, Rouhiainen L, et al. PCR-based identification of microcystin-producing genotypes of different cyanobacterial genera. Arch Microbiol. 2003;180:402-10.
- Ishida K, Christiansen G, Yoshida WY, et al. Biosynthesis and structure of Aeruginoside 126A and 126B Cyanobacterial peptide glycosides bearing a 2-carboxy-6-hydroxyoctahydroindole moiety. Chem Biol. 2007;14:565-76.
- Iteman I, Rippka R, De Marsac NT, et al. Comparison of conserved structural and regulatory domains within divergent 16S rRNA-23S rRNA spacer sequences of cyanobacteria. Microbiol. 2000;146:1275-86.
- Jonasson S, Vintila S, Sivonen K, et al. Expression of the nodularin synthetase genes in the Baltic Sea bloom forming cyanobacterium Nodularia spumigena strain AV1. FEMS Microbiol Ecol. 2008;65:31-9.
- Jungblut AD, Neilan BA. Molecular identification and evolution of the cyclic peptide hepatotoxins microcystin and nodularin synthetase genes in three orders of cyanobacteria. Arch Microbiol. 2006;185:107-14.
- Kellmann R, Michali TK, Neilan BA. Identification of a saxitoxin biosynthesis gene with a history of frequent horizontal gene transfers. J Mol Evol. 2008;67:526-38.
- Koskenniemi K, Lyra C, Rajaniemi-Wacklin P, et al. Quantitative real-time PCR detection of toxic Nodularia cyanobacteria in the Baltic Sea. Appl Environ Microbiol. 2007;73:2173-79.
- Lewis J, Medlin LK, Raine R. MIDTAL (Microarrays for the Detection of Toxic Algae) a Protocol for a Successful Microarray Hybridisation and Analysis Oberreifenberg: Koeltz. 2012.
- Lyra C, Laamanen M, Lehtimäki JM, et al. Benthic cyanobacteria of the genus Nodularia are non-toxic without gas vacuoles able to glide and genetically more diverse than planktonic Nodularia. Int J Syst Evol Microbiol. 2005;55:555-68.
- Mihali TK, Kellmann R, Neilan BA. Characterisation of the paralytic shellfish toxin biosynthesis gene clusters in Anabaena circinalis AWQC131C and Aphanizomenon sp. NH-5. BMC Biochem. 2009;10:8.
- Moffitt M, Blackburn S, Neilan B. rRNA sequences reflect the ecophysiology and define the toxic cyanobacteria of the genus Nodularia. Int J Syst Bacteriol. 2001;51:505-12.
- Moffitt MC, Neilan BA. On the presence of peptide synthetase and polyketide synthase genes in the cyanobacterial genus Nodularia. FEMS Microbiol Lett. 2001;196:207-14.
- Neilan BA, Dittmann E, Rouhiainen L, et al. Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J Bacteriol. 1999;181:4089-97.
- Neilan BA, Jacobs D, Del Dot T, et al. rRNA Sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int J Syst Bacteriol. 1997;47:693-97.
- Noguchi T, Shinohara A, Nishizawa A, et al. Genetic analysis of the microcystin biosynthesis gene cluster in Microcystis strains from four bodies of eutrophic water in Japan. J Gen Appl Microbiol. 2009;55:111-23.
- Nonneman D, Zimba PV. A PCR-based test to assess the potential for microcystin occurrence in channel catfish production ponds. J Phycol. 2002;38;230-33.
- Nübel U, Garcia-Pichel F and Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997;63:3327-32.
- Ostermaier V, Kurmayer R. Application of real-time PCR to estimate toxin production by the cyanobacterium Planktothrix sp. Appl Environ Microbiol. 2010;76:3495-3502.
- Ostermaier V, Kurmayer R. Distribution and abundance of nontoxic mutants of cyanobacteria in lakes of the Alps. Microb Ecol. 2009;58:323-33.
- Ouahid Y, Pirez-Silva G, del Campo FF. Identification of potentially toxic environmental Microcystis by individual and multiple PCR amplification of specific microcystin synthetase gene regions. Environ Toxicol. 2005;20:235-42.
- Rajaniemi P, Hrouzek P, Katovská K, et al. Phylogenetic and morphological evaluation of the genera Anabaena, Aphanizomenon, Trichormus and Nostoc (Nostocales Cyanobacteria). Int Microbiol. 2005;55:11-26.
- Rantala A, Fewer DP, Hisbergues M, et al. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc Natl Acad Sci. 2004;101:568-73.
- Rantala A, Rizzi E, Castiglioni B, et al. Identification of hepatotoxin-producing cyanobacteria by DNA-chip. Environ Microbiol. 2008;10:653-64.
- Rasmussen JP, Giglio S, Monis PT, et al. Development and field testing of a real time PCR assay for cylindrospermopsin producing cyanobacteria. J Appl Microbiol 2008;104:1503-15.
- Rudi K, Skulberg OM, Jakobsen KS. Evolution of cyanobacteria by exchange of genetic material among phyletically related strains. J Bacteriol 1998;180:3453-61.
- Rudi K, Skulberg OM, Skulberg R, et al. Application of sequence-specific labeled 16S rRNA gene oligonucleotide probes for genetic profiling of cyanobacterial abundance and diversity by array hybridization. Appl Environ Microbiol. 2000;66:4004-11.
- Schembri MA, Neilan BA, Saint CP. Identification of genes implicated in toxin production in the cyanobacterium Cylindrospermopsis raciborskii. Environ Toxicol. 2001;16:413-21.
- Schoenhuber W, Zarda B, Eix S, et al. In situ identification of cyanobacteria with horseradish peroxidase-labeled rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1999;65:1259-67.
- Sipari H, Rantala-Ylinen A, Jokela J, et al. Development of a chip assay and quantitative PCR for detecting microcystin synthetase E gene expression. Appl Environ Microbiol. 2010;76:3797-3805.
- Svenning MM, Eriksson T, Rasmussen U. Phylogeny of symbiotic cyanobacteria within the genus Nostoc based on 16S rDNA sequence analyses. Arch Microbiol. 2004;183:19-26.
- Tillett D, Parker DL, Neilan BA. Detection of toxigenicity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis comparison of toxicities with 16S rRNA and phycocyanin operon (Phycocyanin Intergenic Spacer) phylogenies. Appl Environ Microbiol. 2001;67:2810-18.
- Tooming-Klunderud A, Fewer DP, Rohrlack T, et al. Evidence for positive selection acting on microcystin synthetase adenylation domains in three cyanobacterial genera. BMC Evol Biol. 2008;8:256.
- Vaitomaa J, Rantala A, Halinen K, et al. Quantitative real-time PCR for determination of microcystin synthetase e copy numbers for Microcystis and Anabaena in lakes. Appl Environ Microbiol. 2003;69:7289-97.
- Valério E, Chambel L, Paulino S, et al. Molecular identification typing and traceability of cyanobacteria from freshwater reservoirs. Microbiol. 2009;155:642-56.
- Valério E, Pereira P, Saker ML, et al. Molecular characterization of Cylindrospermopsis raciborskii strains isolated from Portuguese freshwaters. Harm Algae. 2005;4:1044-1052.
- Wilson KM, Schembri MA, Baker PD, et al. Molecular characterization of the toxic cyanobacterium Cylindrospermopsis raciborskii and design of a species-specific PCR. Appl Environ Microbiol. 2000;66:332-38.
- Dittami SM, Edvardsen B. GPR-Analyzer A simple tool for quantitative analysis of hierarchical multispecies microarrays. Environ Sci Pollut Res. 2013;20:6808-815.
- Casabiana ML. France-The Mediterraean Lagoons. In: Schrann W. Nienhuis P. Eds, Marine Benthic Vegetation-Recent Changes in and the Effects of Eutrophication. Berlin/Heidelberg. 1966;23:307-29.
- Pacheco AB, Guedes IA, Azevedo SM. Is qPCR a reliable indicator of cyanotoxin risk in freshwater? Toxins. 2016;8:172.
- Warren JA. Review of PCR Inhibition and Its Implications for Human Identity Testing 2013; pp. 1-64.