Establishing a methodology to study the influence of quercetin on the metabolism of the K562/ADM cell line using nuclear magnetic resonance technology
- *Corresponding Author:
- Dr Cai Yu
College of Pharmacy, JiNan University, 601 Huangpu Avenue West, Guangzhou, Guangdon 510632, China; Mr Cai Tiange,China.
Telephone: 86 13119582329
E-mail: caiyu8@sohu.com
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact support@pulsus.com
[ft_below_content] =>Keywords
K562/ADM; Metabolism; Methodology; Quercetin
Metabonomics was initially defined by Nicholson et al (1) in 1999 and subsequently developed into a discipline that quantitatively describes the endogenous metabolites of an organism and its changes (2). Metabonomics research is based on biological fluids, cell extractions, cell culture fluid and tissues, and often use HPLC, GC, MS, NMR, IR and other technologies for detection (3). Quercetin is a natural polyhydroxy flavonoid that commonly exists in fruit, vegetables and other natural plants. It has general pharmacological action, such as antitumour, antioxidant, anti-inflammatory and antithrombotic properties (5,6), and can reverse the multiple drug resistance of leukemia cells (7,8). Metabolites, which are the material foundation of discovering and identifying biomarkers, play an important role in studying cell metabolism. However, to obtain integral metabolites and comprehensive metabolite profiling analyses, optimal extraction methods are needed. The present study established a convenient methodology for isolating metabolites based on researching the influence of quercetin on the metabolism of the drug-resistant leukemia cell line K562/ADM.
| Material, instrumentation and supplier summary |
| SW-CJ-IFD clean bench (Sujing Group, Suzhou Antai Air Technology Co, Ltd); SC-04 low speed centrifuge (Anhui Zhongke Zhongjia Science Instrument Co, Ltd); TS100 inverted fluores- cence microscope (Nikon, edipse); constant temperature carbon dioxide incubator (Thermo Scientific); SB5200D ultrasonic cleaner (Ningbo Xinzhi Biological Polytron Technologies Inc); ACCULAB one over ten-thousand analytical balance (Beijing Sartorius Instrument System Co, Ltd); LS-50HJ vertical pressure steam sterilizer (Jiangyin Binjiang Medical Equipment Co, Ltd); 500 MHz NMR (Bruker, Germany); cell culture flask (America, Corning); dimethyl sulfoxide (Beijing PuBoXin biotechnology co, Ltd); modified RPMI-1640 culture medium (Gibco); Sijiqing fetal bovine serum (Zhejiang Tianhang Biological Technology Co, Ltd); phosphate-buffered saline (Thermo Fisher biochemical products Beijing Co, Ltd, Beijing, China); penicillin and streptomycin solution (100×, Beyotime Institute of Biotechnology); D2O, 3-(trimethylsilyl)propionic acid sodium salt (Shenzhen, Merrill Chemical Technology Co, Ltd); quercetin API (lot number 2013080601, content =97.0%, Guangzhou Aichun Pharmaceutical Science and Technology Co, Ltd); K562/ADM cell line (Nanjing Kaiji Biological Technology Development Co, Ltd). |
Methods
Cell culture
Cells (K562/ADM) were removed from liquid nitrogen storage and quickly placed in a water bath at 37°C for thawing, and subsequently transferred to a centrifuge tube for centrifuging at 1000 rpm for 3 min. The supernatant was removed, and 10% fetal bovine serum (FBS) and 1% (v/v) penicillin/streptomycin RPMI culture solution was added to the resuspended cells. K562/ADM cells were transferred to a cell culture flask and mixed using a pipette, then incubated at 37°C with 5% CO2 at a density of 5×104 cells/mL to 7×l04 cells/mL. Recovered cells were transferred into centrifuge tube and centrifuged at 1000 rpm for 5 min. Following removal of the supernatent, the cells were resuspended in fresh medium and transferred into a culture flask followed by incubation at 37°C with 5% CO2.
Cell passage
Cell passage was performed when cell density reached ≥80%. Cells were transferred to a centrifuge tube at 1000 rpm for 5 min and the supernatent was discarded. Next, the cells were resuspended and washed with phosphate-buffered saline (PBS) solution one to three times. Cells were resuspended in fresh culture medium and transferred into a culture flask at a density of 5×104 cells/mL to 7×l04 cells/mL, then incubated at 37°C with 5% CO2.
Cell cryopreservation
Cell cultures that exhibited good growth were collected in the logarithmic growth phase, centrifuged and counted before cryopreservation . Cell density needed to be controlled at 5×105 cells/mL to 6×105 cells/mL.
Next, cells were placed in cell freezing medium (1:9 ratio of DMSO:FBS) and blended; the solution was then stored at 4°C, −20°C, −80°C for 10 min, 30 min and 16 h to 18 h, respectively. All samples were ultimately placed in liquid nitrogen.
Cell experiment
K562/ADM cells in the logarithmic growth phase were collected, counted and transferred to a 75 cm2 cell culture flask at a density of 1×105 cells/mL. Samples were divided into four groups according to the final concentration of quercetin used in the experiments: blank control (culture solution with 1:1000 DMSO); low dose (10 μmol/L); medium dose (20 μmol/L); and high dose (40 μmol/L). All of the above were incubated at 37°C with 5% CO2.
Cell collection and processing
After 72 h in culture, the cells were centrifuged at 1000 rpm for 5 min and then collected after washing with PBS three times followed by flash freezing in liquid nitrogen. The cells underwent three freeze-thaw cycles before NMR treatment. Two millilitres of 80% methanol was added to the samples and blended, and ultrasonication (3 s on/2 s off) was performed while the samples were on an ice bath for 10 min, followed by centrifugation at 12,000 rpm for 15 min. The supernatant was collected and 1 mL 80% methanol was added to the residue. This treatment was repeated twice. The supernatants were then combined and dried using a nitrogen blow down evaporator.
Collection and processing of cell culture fluid
Cells were centrifuged after culture for 72 h and the supernatant was collected, of which 3 mL was obtained for filtering using a 0.22 μm filter (Waters, USA). Subsequently, centrifugation was performed at 12,000 rpm for 20 min followed by storage in liquid nitrogen. Solvent was removed under vacuum and the residue was freeze-dried to obtain freeze-dried powder before NMR treatment.
NMR sample analysis
Sample preparation: The cell extracts were dissolved in 6 mL 0.01% TMSP/D2O solution and centrifuged at 15,000 rpm for 20 min; 0.5 mL of supernatant was placed in an NMR tube and kept cold for analysis.
NMR detection
All 1H-NMR spectra were generated using 500 MHz NMR (Bruker, Germany) with a 5 mm probe. A standard NOESY impulse sequence was used to obtain spectra of the cell extracts. In this pulse sequence, wait time (RD, 2 s) and mixing time (tm 0.1 s) was conducted water peak suppression. Related parameters were set as follows: SWH, 12 kHz; sampling time, 1.36 s; TD, 32768; NS, 64. All FID signals of 1H-NMR were multiplied by an exponential window function whose broadening factor was 0.5 Hz before Fourier transform.
Results
Morphological observation of K562/ADM cells
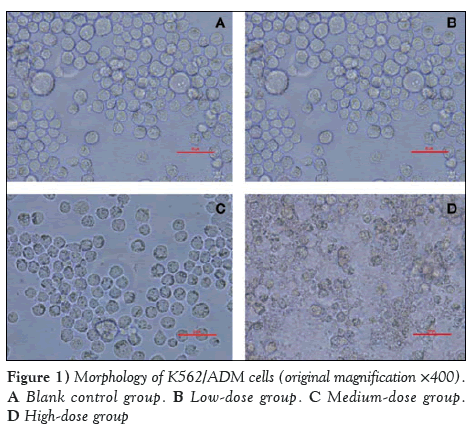
Cellular morphology of the K562/ADM cells after treatment with quercetin for 72 h was assessed under reverse microscopy (original magnification ×400) (Figure 1).
As shown in Figure 1, the cellular shape of the blank control group (1:1000 DMSO) is high-blooded without shrinkage or cracking. No significant difference was observed compared with pretherapy. However, in the low-dose group (10 μmol/L quercetin), a small number of cells had ruptured and exhibited lower activity. In the high-dose group (40 μmol/L quercetin), the phenomenon of cell shrinkage and rupture was obvious. Thus, it was educible that quercetin could inhibit the growth of K562/ADM in a dose-dependent manner.
NMR assessment of cell extracts
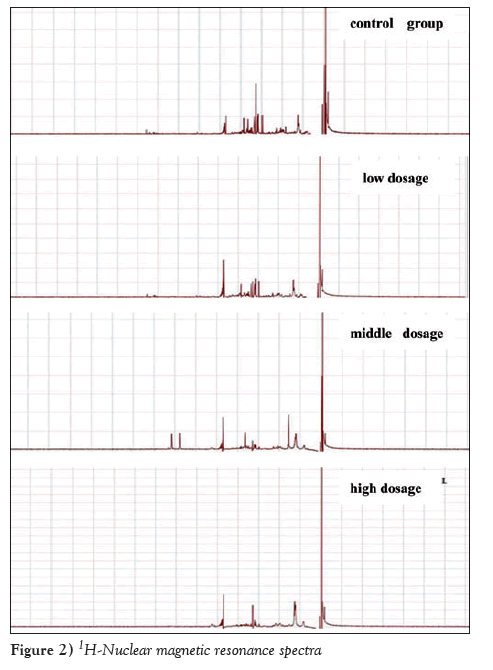
NMR assessment was conducted as described, and the results obtained underwent Fourier transform to obtain 1H-NMR spectra, which were then conducted phase calibration and baseline adjustment. Chemical displacement of all spectra was calibrated with TMSP (δ=0.00). TMSP-marked peaks were obvious in the four 1H-NMR spectra. There was apparent characteristic peak and high peak value, as shown in Figure 2.
Discussion
Drug-resistant K562/ADM cells were cultured in 1000 ng/mL adriamycin medium to maintain drug resistance (9). To avoid the influence of adriamycin on the results, K562/ADM cells should be cultured in a normal culture medium without adriamycin for two weeks before the experiment. Two weeks later, K562/ADM cells should be placed into culture solution containing adriamycin so as to maintain drug resistance.The process of cell collection and processing was implemented after treatment with quercetin for 72 h. The present study vacuum freeze-dried the cell extracts by repeated freezing and thawing the cell extract three times to inactivate the cells, then an 80% methanol solution was added to extract the intracellular metabolites; the collected metabolite solutions were subsequently freeze dried to powder form. However, this method required a lyophilizer to remove organic solvent, which required specialized equipment and not easy to conduct. Therefore, a nitrogen blow down evaporator was ultimately chosen for drying the samples.
There have been many detection technologies used in metabonomics research includiung HPLC, MS and GC. The present study chose NMR for the following advantages (10-12): Pre-treatment of samples was easy and the required detection amount was small.
Detection could be performed as soon as D2O was added for field lock and special treatment of the samples was unnecessary. The structure and original properties of the samples were not destroyed; detection time is short (approximately 5 min to 10 min); and sample purification was not necessary. Meanwhile, various two-dimensional spectroscopy could be used for the determination and structural identification of unknown metabolites.
Disclosures
The authors have no financial disclosures or conflicts of interest to declare.
Foundation Support
The General Program of the National Natural Science Foundation of China (NO. 81173215). The ministry of education in the new century excellent talents to support plan (NO. NECT-12-0677). Guangzhou science and technology cooperation platform project (2014J4500005).
References
- Nicholson JK, Lindon JC, Holmes E. ?Metabonomics?:understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999;29:1181-9.
- Tang HR, Wang YL. Metabonomics: A revolution in progress. Prog Biochem Biophys 2006;33:401-17.
- Lenz EM, Wilson ID. Analytical strategies in metabonomics. J Proteome Res 2007;6:443-58.
- Sun J, Yu S. Research progress of quercetin. Research and Practice on Chinese Medicines 2011;3:85-8.
- Han Y, Cao L, Hao H, et al. Study of quercetin reverses the expression of multidrug resistance in leukemia cell line K562/A and the related genes. J Exp Hematol 2011;4:884-9.
- Liu M, Xiao Y, Zuo A, et al. The research of Quercetin, phloretin, silybin scavenge free radicals and inhibit lipid peroxidation activity. Chinese patent medicine 2012;4:753-6.
- Shu Y, Tan T, Zhang S, et al. Progress in pharmacological research of quercetin. West China J Pharmaceutical Sci 2008;6:689-91.
- Chen C, Cai T, Zhang R, et al. Effects of 6 kinds of active ingredients of Chinese herbal medicine on reversing drug resistance of K562/ADM and P27 , P170 protein expression. Chinese Patent Medicine 2014;1:1-4.
- Gai X, Li C, Li Q, et al. Reversal effect of saikosaponin on human leukemia cell line K562/ADM in multidrug resistance in vitro. Chin J Pathophysiol 2012;1:76-80.
- Wang weiguang,Yu guangqing,Zhang guoyan,et al. Quercetin reverses multidrug resistance on K562/ADM in vitro. Heilongjiang Medicine and Pharmacy 2011;34:28-9.
- Miao C,Wang Y. Application of NMR technique in the discovery and pharmacological studies of active substances from natural products. Acta Pharmaceutica Sinica 2013;48:1383-9.
- Zou Y, He J, Xie H,et al. Application of metabolic studies in animal nutrition on the basis of NMR technology.Chin J Animal Nutr 2012;24:2073-8.
- *Corresponding Author:
- Dr Cai Yu
College of Pharmacy, JiNan University, 601 Huangpu Avenue West, Guangzhou, Guangdon 510632, China; Mr Cai Tiange,China.
Telephone: 86 13119582329
E-mail: caiyu8@sohu.com
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact support@pulsus.com
Abstract
The purpose of the present study was to establish an experimental method to study the effects of quercetin on the metabolism of the drug-resistant leukemia cell line K562/ADM using nuclear magnetic resonance (NMR) technology. Drug administration to the cells was performed according to proper experimental design and application. The cells were collected and inactivated by repeated freeze-thaw cycles using liquid nitrogen. Methanol (80%) was used to extract cell metabolites, and a sample concentrator (Termovap, ECOM, Czech Republic) was used to purify the samples. All samples were assessed using 500 MHz NMR (Bruker, Germany) with standard NOESY impulse sequence to obtain 1H-NMR spectra of cell metabolites. The processing method used to assess cell metabolism established in the present study was convenient and led to minimal loss of cell metabolites. The 1H-NMR spectra obtained using NMR technology reliably reflected changes in cell metabolism.
-Keywords
K562/ADM; Metabolism; Methodology; Quercetin
Metabonomics was initially defined by Nicholson et al (1) in 1999 and subsequently developed into a discipline that quantitatively describes the endogenous metabolites of an organism and its changes (2). Metabonomics research is based on biological fluids, cell extractions, cell culture fluid and tissues, and often use HPLC, GC, MS, NMR, IR and other technologies for detection (3). Quercetin is a natural polyhydroxy flavonoid that commonly exists in fruit, vegetables and other natural plants. It has general pharmacological action, such as antitumour, antioxidant, anti-inflammatory and antithrombotic properties (5,6), and can reverse the multiple drug resistance of leukemia cells (7,8). Metabolites, which are the material foundation of discovering and identifying biomarkers, play an important role in studying cell metabolism. However, to obtain integral metabolites and comprehensive metabolite profiling analyses, optimal extraction methods are needed. The present study established a convenient methodology for isolating metabolites based on researching the influence of quercetin on the metabolism of the drug-resistant leukemia cell line K562/ADM.
| Material, instrumentation and supplier summary |
| SW-CJ-IFD clean bench (Sujing Group, Suzhou Antai Air Technology Co, Ltd); SC-04 low speed centrifuge (Anhui Zhongke Zhongjia Science Instrument Co, Ltd); TS100 inverted fluores- cence microscope (Nikon, edipse); constant temperature carbon dioxide incubator (Thermo Scientific); SB5200D ultrasonic cleaner (Ningbo Xinzhi Biological Polytron Technologies Inc); ACCULAB one over ten-thousand analytical balance (Beijing Sartorius Instrument System Co, Ltd); LS-50HJ vertical pressure steam sterilizer (Jiangyin Binjiang Medical Equipment Co, Ltd); 500 MHz NMR (Bruker, Germany); cell culture flask (America, Corning); dimethyl sulfoxide (Beijing PuBoXin biotechnology co, Ltd); modified RPMI-1640 culture medium (Gibco); Sijiqing fetal bovine serum (Zhejiang Tianhang Biological Technology Co, Ltd); phosphate-buffered saline (Thermo Fisher biochemical products Beijing Co, Ltd, Beijing, China); penicillin and streptomycin solution (100×, Beyotime Institute of Biotechnology); D2O, 3-(trimethylsilyl)propionic acid sodium salt (Shenzhen, Merrill Chemical Technology Co, Ltd); quercetin API (lot number 2013080601, content =97.0%, Guangzhou Aichun Pharmaceutical Science and Technology Co, Ltd); K562/ADM cell line (Nanjing Kaiji Biological Technology Development Co, Ltd). |
Methods
Cell culture
Cells (K562/ADM) were removed from liquid nitrogen storage and quickly placed in a water bath at 37°C for thawing, and subsequently transferred to a centrifuge tube for centrifuging at 1000 rpm for 3 min. The supernatant was removed, and 10% fetal bovine serum (FBS) and 1% (v/v) penicillin/streptomycin RPMI culture solution was added to the resuspended cells. K562/ADM cells were transferred to a cell culture flask and mixed using a pipette, then incubated at 37°C with 5% CO2 at a density of 5×104 cells/mL to 7×l04 cells/mL. Recovered cells were transferred into centrifuge tube and centrifuged at 1000 rpm for 5 min. Following removal of the supernatent, the cells were resuspended in fresh medium and transferred into a culture flask followed by incubation at 37°C with 5% CO2.
Cell passage
Cell passage was performed when cell density reached ≥80%. Cells were transferred to a centrifuge tube at 1000 rpm for 5 min and the supernatent was discarded. Next, the cells were resuspended and washed with phosphate-buffered saline (PBS) solution one to three times. Cells were resuspended in fresh culture medium and transferred into a culture flask at a density of 5×104 cells/mL to 7×l04 cells/mL, then incubated at 37°C with 5% CO2.
Cell cryopreservation
Cell cultures that exhibited good growth were collected in the logarithmic growth phase, centrifuged and counted before cryopreservation . Cell density needed to be controlled at 5×105 cells/mL to 6×105 cells/mL.
Next, cells were placed in cell freezing medium (1:9 ratio of DMSO:FBS) and blended; the solution was then stored at 4°C, −20°C, −80°C for 10 min, 30 min and 16 h to 18 h, respectively. All samples were ultimately placed in liquid nitrogen.
Cell experiment
K562/ADM cells in the logarithmic growth phase were collected, counted and transferred to a 75 cm2 cell culture flask at a density of 1×105 cells/mL. Samples were divided into four groups according to the final concentration of quercetin used in the experiments: blank control (culture solution with 1:1000 DMSO); low dose (10 μmol/L); medium dose (20 μmol/L); and high dose (40 μmol/L). All of the above were incubated at 37°C with 5% CO2.
Cell collection and processing
After 72 h in culture, the cells were centrifuged at 1000 rpm for 5 min and then collected after washing with PBS three times followed by flash freezing in liquid nitrogen. The cells underwent three freeze-thaw cycles before NMR treatment. Two millilitres of 80% methanol was added to the samples and blended, and ultrasonication (3 s on/2 s off) was performed while the samples were on an ice bath for 10 min, followed by centrifugation at 12,000 rpm for 15 min. The supernatant was collected and 1 mL 80% methanol was added to the residue. This treatment was repeated twice. The supernatants were then combined and dried using a nitrogen blow down evaporator.
Collection and processing of cell culture fluid
Cells were centrifuged after culture for 72 h and the supernatant was collected, of which 3 mL was obtained for filtering using a 0.22 μm filter (Waters, USA). Subsequently, centrifugation was performed at 12,000 rpm for 20 min followed by storage in liquid nitrogen. Solvent was removed under vacuum and the residue was freeze-dried to obtain freeze-dried powder before NMR treatment.
NMR sample analysis
Sample preparation: The cell extracts were dissolved in 6 mL 0.01% TMSP/D2O solution and centrifuged at 15,000 rpm for 20 min; 0.5 mL of supernatant was placed in an NMR tube and kept cold for analysis.
NMR detection
All 1H-NMR spectra were generated using 500 MHz NMR (Bruker, Germany) with a 5 mm probe. A standard NOESY impulse sequence was used to obtain spectra of the cell extracts. In this pulse sequence, wait time (RD, 2 s) and mixing time (tm 0.1 s) was conducted water peak suppression. Related parameters were set as follows: SWH, 12 kHz; sampling time, 1.36 s; TD, 32768; NS, 64. All FID signals of 1H-NMR were multiplied by an exponential window function whose broadening factor was 0.5 Hz before Fourier transform.
Results
Morphological observation of K562/ADM cells
Cellular morphology of the K562/ADM cells after treatment with quercetin for 72 h was assessed under reverse microscopy (original magnification ×400) (Figure 1).
Figure 1: Morphology of K562/ADM cells (original magnification ×400). A Blank control group. B Low-dose group. C Medium-dose group. D High-dose group
As shown in Figure 1, the cellular shape of the blank control group (1:1000 DMSO) is high-blooded without shrinkage or cracking. No significant difference was observed compared with pretherapy. However, in the low-dose group (10 μmol/L quercetin), a small number of cells had ruptured and exhibited lower activity. In the high-dose group (40 μmol/L quercetin), the phenomenon of cell shrinkage and rupture was obvious. Thus, it was educible that quercetin could inhibit the growth of K562/ADM in a dose-dependent manner.
NMR assessment of cell extracts
NMR assessment was conducted as described, and the results obtained underwent Fourier transform to obtain 1H-NMR spectra, which were then conducted phase calibration and baseline adjustment. Chemical displacement of all spectra was calibrated with TMSP (δ=0.00). TMSP-marked peaks were obvious in the four 1H-NMR spectra. There was apparent characteristic peak and high peak value, as shown in Figure 2.
Figure 2: 1H-Nuclear magnetic resonance spectra
Discussion
Drug-resistant K562/ADM cells were cultured in 1000 ng/mL adriamycin medium to maintain drug resistance (9). To avoid the influence of adriamycin on the results, K562/ADM cells should be cultured in a normal culture medium without adriamycin for two weeks before the experiment. Two weeks later, K562/ADM cells should be placed into culture solution containing adriamycin so as to maintain drug resistance.The process of cell collection and processing was implemented after treatment with quercetin for 72 h. The present study vacuum freeze-dried the cell extracts by repeated freezing and thawing the cell extract three times to inactivate the cells, then an 80% methanol solution was added to extract the intracellular metabolites; the collected metabolite solutions were subsequently freeze dried to powder form. However, this method required a lyophilizer to remove organic solvent, which required specialized equipment and not easy to conduct. Therefore, a nitrogen blow down evaporator was ultimately chosen for drying the samples.
There have been many detection technologies used in metabonomics research includiung HPLC, MS and GC. The present study chose NMR for the following advantages (10-12): Pre-treatment of samples was easy and the required detection amount was small.
Detection could be performed as soon as D2O was added for field lock and special treatment of the samples was unnecessary. The structure and original properties of the samples were not destroyed; detection time is short (approximately 5 min to 10 min); and sample purification was not necessary. Meanwhile, various two-dimensional spectroscopy could be used for the determination and structural identification of unknown metabolites.
Disclosures
The authors have no financial disclosures or conflicts of interest to declare.
Foundation Support
The General Program of the National Natural Science Foundation of China (NO. 81173215). The ministry of education in the new century excellent talents to support plan (NO. NECT-12-0677). Guangzhou science and technology cooperation platform project (2014J4500005).
References
- Nicholson JK, Lindon JC, Holmes E. ?Metabonomics?:understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999;29:1181-9.
- Tang HR, Wang YL. Metabonomics: A revolution in progress. Prog Biochem Biophys 2006;33:401-17.
- Lenz EM, Wilson ID. Analytical strategies in metabonomics. J Proteome Res 2007;6:443-58.
- Sun J, Yu S. Research progress of quercetin. Research and Practice on Chinese Medicines 2011;3:85-8.
- Han Y, Cao L, Hao H, et al. Study of quercetin reverses the expression of multidrug resistance in leukemia cell line K562/A and the related genes. J Exp Hematol 2011;4:884-9.
- Liu M, Xiao Y, Zuo A, et al. The research of Quercetin, phloretin, silybin scavenge free radicals and inhibit lipid peroxidation activity. Chinese patent medicine 2012;4:753-6.
- Shu Y, Tan T, Zhang S, et al. Progress in pharmacological research of quercetin. West China J Pharmaceutical Sci 2008;6:689-91.
- Chen C, Cai T, Zhang R, et al. Effects of 6 kinds of active ingredients of Chinese herbal medicine on reversing drug resistance of K562/ADM and P27 , P170 protein expression. Chinese Patent Medicine 2014;1:1-4.
- Gai X, Li C, Li Q, et al. Reversal effect of saikosaponin on human leukemia cell line K562/ADM in multidrug resistance in vitro. Chin J Pathophysiol 2012;1:76-80.
- Wang weiguang,Yu guangqing,Zhang guoyan,et al. Quercetin reverses multidrug resistance on K562/ADM in vitro. Heilongjiang Medicine and Pharmacy 2011;34:28-9.
- Miao C,Wang Y. Application of NMR technique in the discovery and pharmacological studies of active substances from natural products. Acta Pharmaceutica Sinica 2013;48:1383-9.
- Zou Y, He J, Xie H,et al. Application of metabolic studies in animal nutrition on the basis of NMR technology.Chin J Animal Nutr 2012;24:2073-8.