Generation of blaNDM-1 super MDR gene by multiple rearrangement of other metallo-beta-lactamases like GIM-1, IMP-1 and VIM-2 including many IS-elements
Received: 04-Jan-2018 Accepted Date: Jan 30, 2018; Published: 07-Feb-2018
Citation: Chakraborty AK, Nandi SK. Generation of blaNDM-1 super MDR gene by multiple rearrangement of other metallo-beta-lactamases like GIM-1, IMP-1 and VIM-2 including many IS-elements. J Microbiol Biotechnol Rep. 2018;2(1):17-21.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Discovery of blaNDM-1 β-lactamase in plasmids of Escherichia coli and Pseudomonas aeruginosa and unresponsiveness of such infections to most antibiotics including imipenem and dorripenem shocked the whole drug industry in 2009 as well as discovery of mcr-1 gene in 2016 orchested such horror being colistin drug was useless now. We have checked such horror in Ganga River water which contains superbugs, resistant to most common antibiotics including methicillin, lomofloxacin, amikacin, imipenem, linozolid and vancomycin. Beta-lactamases (TEM, OXA, CTX-M, CMY, GIM, VIM, NDM, KPC), drug modifying acetyl/phospho/adenyl transferases and drugs efflux proteins (tetA/C, mexAB/CD/EF) are mostly diversified and mutated creating hundred to thousand isomers. Many MDR genes are accumulated now in large MDR conjugative plasmids and it is almost impossible to PCR identify all MDR genes in any clinical isolate for better prognosis and therapeutics. From in silico protein alignment studies, we concluded that blaNDM-1 gene might be originated from the rearrangement of other Metallo-β-lactamases genes present in same plasmids or multiple integrons. This conclusion supports our previous studies with Kolkata superbugs Escherichia coli KT-1_ MDR and Pseudomonas aeruginosa DB-2_ MDR which contain multiple small as well as large plasmids with many diverged MDR genes and transposons. Presence of ThiX gene (in pKOX_R1 plasmid) involved in Thiamine biosynthesis suggests the bacteria need vitamin synthesizing enzymes in plasmid if we continue high dose complex antibiotic intake. Thus many IS-elements and transposons are assembled in MDR plasmids for new MDR gene creation to save symbiotic relation between gut microbiota and human metabolosome.
Introduction
β-lactamases have been grouped into four major classes (A–D) based on sequence homology [1] but diverged more profoundly as described recently [2-4]. Classes A, C, and D use an active site serine as a nucleophile. However, the class B metallo-β-lactamase (MBL) enzymes use bound zinc atoms at the active site [5]. Millions of MDR genes have been sequenced that includes mostly bla genes (TEM, SHV, CTX-M-1/2/9, OXA2/23/48/58, VIM, CMY, GIM, VIM etc.), drug modifying genes like catB3, aadA1, aacII-a, aph-Ib and diverse range of drug transporter genes like acrAB, macA/B, mexCD/EF, bcr, bmr, tetA/B, mcr, norA and mtrCDE [6] as well as arr3, sul1/2 and mcr-1. Drug industry is always run to discover new antibiotic derivatives to overcome the actions of MDR genes located in plasmids as well as chromosome. As for example, cefotaxime is good in case of blaTEM containing plasmids but blaCTX-M enzyme lyses cefotaxime and new imipenem drug is introduced. Soon, blaVIM or blaIMP enzymes are developed that destroy imipenem and recently (2009) discovered blaNDM1 could destroy all penicillins, cefallosporins and carbapenems poising a threat to drug industry when no one want to invest in new drug discovery. Lastly, we introduced β-lactamase inhibitor (cavulinate, sulbactam, avibactam) combination therapy but problems continued. Most devastating fact, carbapenemsaes are clustered in large conjugative plasmids in presence of ESBL enzymes like KPC-2, VIM, OXA-23, OXA-210 and diverged drug acetyl/phospho transferases including PBPs and many drug efflux genes [6]. Those bacteria are named as superbugs and such infections must be treated in the developed countries like UK and USA [7-9]. We are now studying the mutational profiles of bla genes in the isolated MDR-bacteria from Ganga River and Rain water of Kolkata [in press].
K. pneumoniae 151kb plasmid (pKP048) (accession no. FJ628167) contains MDR genes like macrolide ABC transporter, floquinolone resistant gene (qnrB4) , mercury resistant gene (MerE), sulphonamide resistant protein (sul1) and aph gene for aminoglycoside phosphotransferases [10]. BlaKPC1 gene was located in K. pneumoniae plasmids (accession nos. NC_022078, NC_014312, JX283456 and KF954759) and blaNDM1 gene was also located in many large conjugative plasmids (CP009116, JN420336 and AP012055) [11]. Plasmid pKOX_R1 contains metal resistant genes and many ABC, MFS, AAA drug transporters as well as common MDR genes like cat, sul1, aac3’-IId, FosA3, ANT, aph and NDM-1, CTX-M-3 and SHV-12 β-lactamases. Most importantly such plasmids has less Tra genes but 16 types of inserted IS-elements and transposons. We investigated the nature of NDM-1 gene creation here as previously we have detected a homology between VIM-2 and NDM-1 [2-4]. We conclude that a critical message has indeed created in bacteria and gut luminal cells to preserve symbiotic relation by continuing synthesis of new MDR genes and acquiring other genetic changes. IS elements, transposases, integrases, recombinases are highly assembled in MDR plasmids and may contribute to the efficient MDR gene creation and AMR void. Interestingly, blaGIM, blaVIM and blaIMP metallo β-lactamases may act as progenitor of super MDR Gene, blaNDM-1.
Materials and Methods
NCBI databases were retrieved using the BLAST programmes (www.ncbi. nlm.nih.gov/blast). The complete genes of blaGIM, blaVIM, blaIMP or blaNDM1 were sequenced in plasmids and were analyzed by Seq- 2 programme of BLAST (NDM-1 Protein Ids. AGL09203; AEA41876). Multalin protein sequence software was used to get the nature of conserved sequences among metallo-class B β-lactamases [12]. Sometime, diverged sequences are manually cut and paste into align position in MS word so that it is appeared both sequences have similarity. For retrieving any nucleotide like blaGIM-1 or blaVIM, we type the same at the NCBI port (www.ncbi.nlm. nih.gov/nucleotide or Protein) and to BL AST search to type the accession number for protein or DNA into BLAST port (https://blast.ncbi.nlm.nih. gov/Blast.cgi?PAGE_TYPE=BlastSearch) [13,14].
Result and Discussion
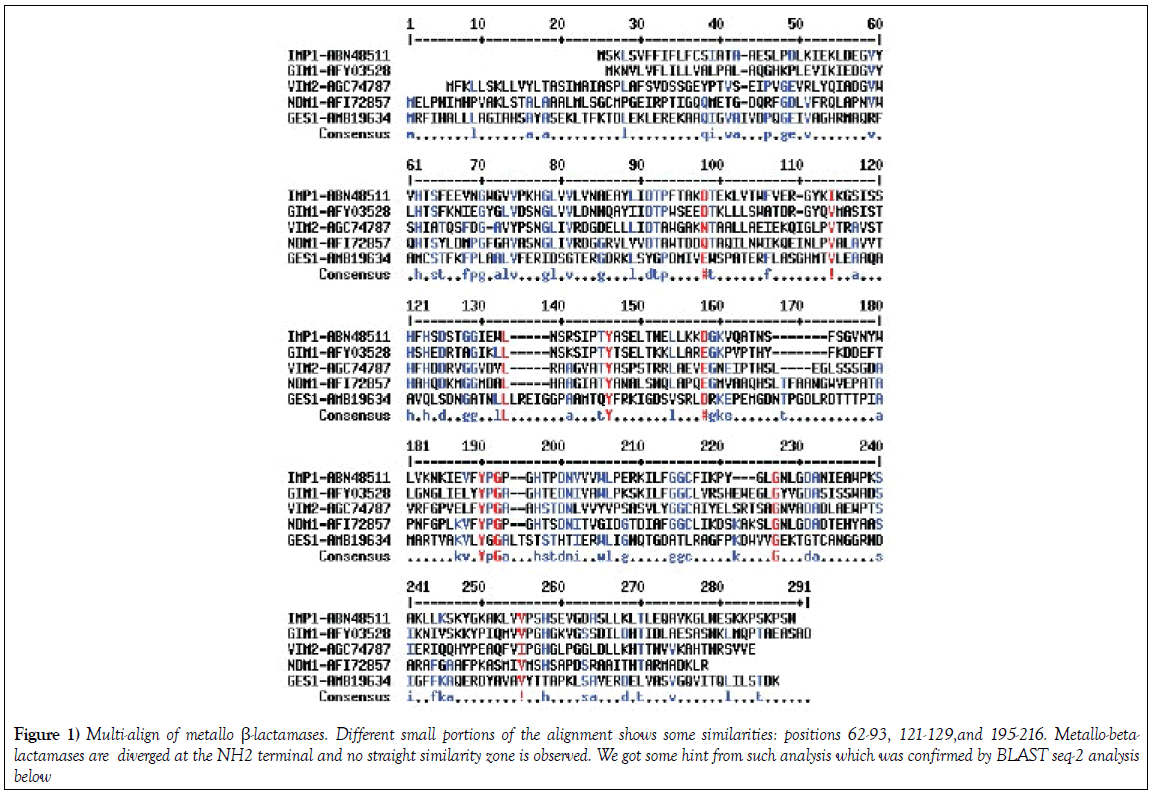
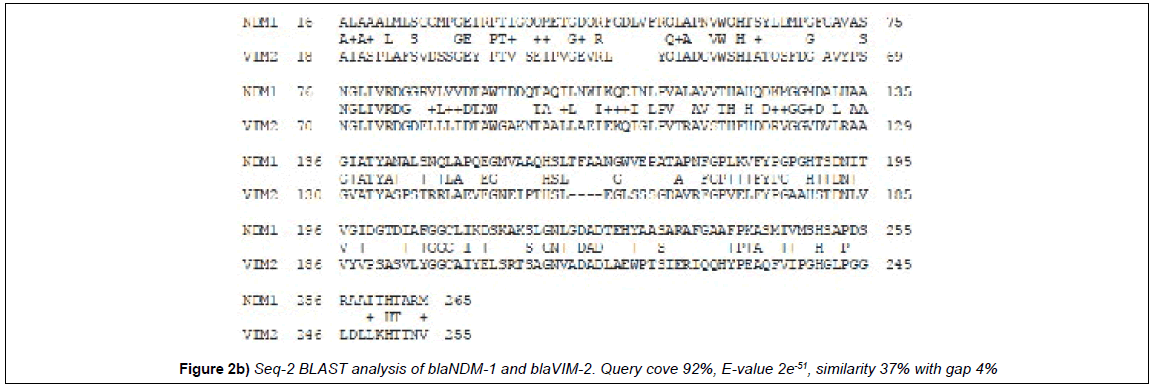
Multiple align data suggests that many MBL β-lactamases have some interesting similarities to the blaNDM-1 protein that degrades all penicillin, cephalosporin and carbapenem drugs (Figure 1). blaNDM-1 and blaKPC-2 containing plasmids are emerging in India and such superbugs are very deadly as unresponsive to most antibiotics [15-17]. We used seq-2 BLAST (www.ncbi.nlm.hih.gov/blast /nucore/protein) analysis to pinpoint the ho,ology between GIM-1, VIM-2 and IMP-1 β-lactamases. NDM-1 at position ‘74’ and VIM-2 at position ‘68’ have “SNGLIVRDG” nine amino acid similarities and at 110 position within 31 amino acids 20 are identical (65%) suggesting blaNDM-1 gene has partially originated from blaVIM-2 gene (Figure 1; Figure 2a). Where as at 62 aa position of NDM1, we see a conserved motifs (HTSYLDMPGFGAVASNGLIRD) with GIM1 protein sequence with 12aa identity (Figure 2b). The seq-2 align suggests that “ VFYPGPGHT” nine amino acids and “LGNLGDA” seven amino acids perfect matches with IMP-1 β-lactamase at 182/218 positions respectively (Figure 2c). and is very interesting as many plasmids carry multiple such genes and may facilitate the generation of blaNDM-1 super protein that destroys all penicillins, cephalosporins and carbapenems [4]. In history of hundreds of MDR genes, NDM-1 containing superbugs have created the most horror in society other than drug modifying (cat, aac, aph) and drug efflux genes (mex and tet). In truth, blaNDM1 and KPC2 containing superbugs are increasing in the environment of India [2,9] and many NDM1 mutants have now sequenced [4]. When we aligned with KPC2 protein sequence and only “156-AAQHSLTFAAN” was aligned considerably (125-AVQYSDNAAAN) suggesting KPC-2 was not involved in that case [18].
Figure 1: Multi-align of metallo β-lactamases. Different small portions of the alignment shows some similarities: positions 62-93, 121-129,and 195-216. Metallo-betalactamases are diverged at the NH2 terminal and no straight similarity zone is observed. We got some hint from such analysis which was confirmed by BLAST seq-2 analysis below
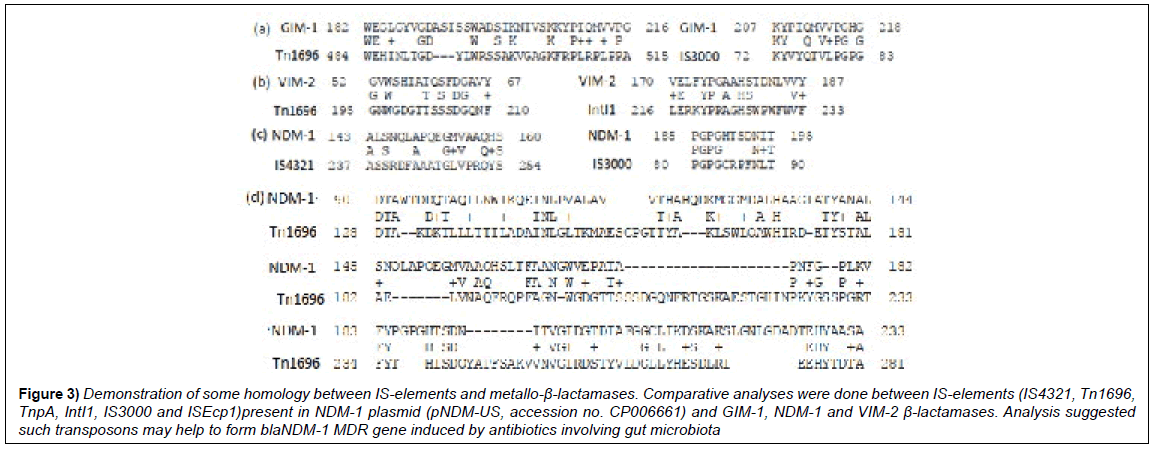
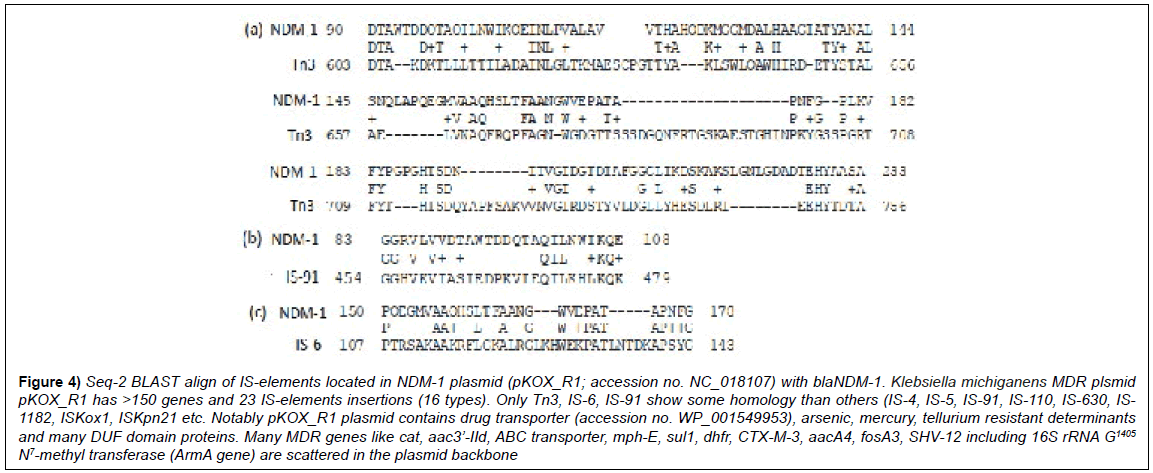
We further investigated the occurrence of transposons and IS-elements in large as well as small MDR plasmids. Many medium sized plasmids (25-40kb) do contains 4-8 MDR genes and few IS-elements containing transposases, resolvases and integrases [3]. We detected that Tn1696, IntI1 and IS3000 have some similarity zones to blaNDM-1 and also to blaGIM-1 (Figure 3). In pKOX_R1 plasmid we found 23 IS insertions and such astonishing result we explored further. Seq-2 BLAST analysis detected few stretch of 30-50% homology with blaNDM-1 protein sequence with Tn3, IS-6 and IS-91 sequences (Figure 4). Small plasmids and integrons (see, accession nos. EF375699; KT984195; JX566715; GQ466184; EU597467; DQ310703; NC_019081) carry VIM-1, GES-1, GIM-1, IMP-1 carbapenemases including ESBL OXA β-lactamases including many integrases and recombinases. Thus, large plasmid (accession nos. AP012055, NC_018107, KT185451) may generate later and blaNDM-1 gene may be generated in small plasmid which directly combined with conjugative plasmid [10]. As for example, isolated imipenem resistant microbes from Ganga River water seem lacking tetracycline, streptomycin and chloramphenicol resistant determinants (unpublished). In truth, imipenem resistant superbugs are rare in the environment (0.002%), although in clinical isolates 5-10% imipenem resistant bacteria are detected. Klebsiella pneumonia large plasmid pCR14_2 (accession no. NZ_CP015394) also have inserted many transposons (IS110, IS91, Tn3, IS5, IntI1) including clustered MDR genes OXA-2, sul1, CTX-M2 and AAC(3’)-IIa.
Figure 3: Demonstration of some homology between IS-elements and metallo-β-lactamases. Comparative analyses were done between IS-elements (IS4321, Tn1696, TnpA, IntI1, IS3000 and ISEcp1)present in NDM-1 plasmid (pNDM-US, accession no. CP006661) and GIM-1, NDM-1 and VIM-2 β-lactamases. Analysis suggested such transposons may help to form blaNDM-1 MDR gene induced by antibiotics involving gut microbiota
Figure 4: Seq-2 BLAST align of IS-elements located in NDM-1 plasmid (pKOX_R1; accession no. NC_018107) with blaNDM-1. Klebsiella michiganens MDR plsmid pKOX_R1 has >150 genes and 23 IS-elements insertions (16 types). Only Tn3, IS-6, IS-91 show some homology than others (IS-4, IS-5, IS-91, IS-110, IS-630, IS- 1182, ISKox1, ISKpn21 etc. Notably pKOX_R1 plasmid contains drug transporter (accession no. WP_001549953), arsenic, mercury, tellurium resistant determinants and many DUF domain proteins. Many MDR genes like cat, aac3’-IId, ABC transporter, mph-E, sul1, dhfr, CTX-M-3, aacA4, fosA3, SHV-12 including 16S rRNA G1405 N7-methyl transferase (ArmA gene) are scattered in the plasmid backbone
Conclusion
MDR horror has reached every home of this Earth and WHO advised new interventions to stop this medical catastrophe [11,19-22]. We found that super conjugative plasmids have many MDR genes in single plasmid and also many small plasmids like integrons and IS-elements [23,24]. We postulated that a critical message has generated between bacteria and human at the intestine to preserve symbiotic relation in presence of high dose of antibiotics that all are taking since 1940s [25]. One high dose of antibiotic kills gut microbiota that synthesize 20 vitamins and complex bio-molecules which are absolutely needed for normal human metabolosome evolving >30000 enzymatic bio-conversions. MDR plasmids contain many transposes, resolvases, topoisomerases and integrases and such enzymes are constantly rearranging genes in bacterial cytoplasm [2,12]. Thus bacteria can make a new MDR gene against new antibiotic in the intestine activated by antibiotic itself and probably within few weeks to save vitamin biosynthesis for human. We proved here that deadly NDM-1 gene is created from plasmids containing blaGIM-1, blaVIM-2 and blaIMP-1 like genes which present in combination with many other genes and IS-elements. We found ThiF gene (in pKOX_R1 plasmid) that involved in thiamine vitamin biosynthesis indicating MDR bacteria will be acquired all vitamin synthesizing enzymes in MDR plasmids gradually if we will be continued complex antibiotic intake. Such large plasmid has few Tra conjugative genes (conjugative deficient) but contains 23 insertion elements suggesting a great site for MDR gene creation. We believe interventions targeting DNA rearrangement in superbugs may be a alternative target to control superbug spread [26-30].
Acknowledgement
We thank Prof. Chandradeepa Ghosh, Department of Human Physiology, Vidyasagar University for help during the study and Dr. J B Medda of Oriental Association of Education and Research for financial help.
REFERENCES
- Ambler RP, Coulson AF, Frere JM, et al. A standard numbering scheme for the class A beta-lactamases. Biochemical J. 1991;276 (Pt 1):269-70.
- Chakraborty AK. Multi-drug resistant genes in bacteria and 21st Century problems associated with antibiotic therapy. Biotechnol Ind J. 2016a;12(12):114.
- ChakrabortyAK. Complexity heterogeneity 3-D structures and transcriptional activation of multi-drug resistant clinically relevant bacterial beta-lactamases. Trends Biotechnol: Open Access. 2016b;2(1):1-001.
- Chakraborty AK. In silico analysis of multidrug resistant bacterial KPC-1 and NDM-1 beta-lactamases that confers severe MDR phenotypes. Biochem Biotechnol Res. 2016c;4(1):17-26.
- Bebrone C. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochemical Pharmacol. 2007;74(12):1686-1701.
- Sun J, Deng Z, Yan A. Bacterial multidrug efflux pumps: Mechanisms physiology and pharmacological exploitation. Biochem Biophys Res Communi. 2014;453:254-67.
- Lynch JP, Clark NM, Zhane GG. Evolution of antimicrobial resistance among Enterobacteriaceae (focus on extended spectrum beta-lactamases and carbapenemases). Exp Opini Pharmacother. 2013;14:199-210.
- Wang L, Fang H, Feng J et al. Complete sequences of KPC-2 encoding p628-KPC and CTX-M-65 encoding p628-CTXM coexisted in Klebsiella kneumoniae. Frontier Microbiol. 2015;6:838.
- Chakraborty AK, Maity M, Patra S, et al. Complexity heterogeneity and mutational analysis of antibiotic inactivating acetyl transferases in MDR conjugative plasmids conferring multi-resistance. Res Rev J Microbiol Biotechnol. 2017;6(2):28-44.
- Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227-38.
- Dotson GA, NISC Comparative Sequencing Program, Dekker JP et al. Draft Genome Sequence of a Klebsiella pneumoniae Carbapenemase-Positive Sequence Type 111 Pseudomonas aeruginosa Strain. Genome Announc. 2016;4(1):e01663-15.
- King DT, Strynadka NC. Targeting metallo-beta-lactamase enzymes in antibiotic resistance. Future Med Chem. 2013;5:1243-63.
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45-8.
- Altschul SF, Madden TL, Schaffer AA et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res.1997;25:3389-3402.
- Ho PL, Lo WU, Yeung MK, et al. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS ONE. 2011;6(3):E17989.
- Kumarasamy K, Kalyanasundaram A. Emergence of Klebsiella pneumoniae isolate co-producing NDM-1 with KPC-2 from India. J Antimicrob Chemother. 2012;67:243-44.
- Paul D, Dhar D, Maurya AP et al. Co-carriage of blaKPC-2 and blaNDM-1 in Clinical Isolates of Pseudomonas aeruginosa associated with Hospital infections from India. PLoS ONE. 2015;10(12):e0145823.
- Campos JC, da Silva MJ, dos Santos PR, et al. Characterization of Tn3000 a transposon responsible for blaNDM-1 dissemination among Enterobacteriaceae in Brazil, Nepal, Morocco and India. Antimicrob Agents Chemother. 2015;59:7387-95.
- Chakraborty AK. Mechanisms of AMR: Bacteria Won the Battle Against Antibiotics. Insights Biomed. 2017b;2(4):18.
- Chakraborty AK. Colistin drug resistant determinant Mcr-1 gene spreads in conjugative plasmids creating huge confusion for the treatment of multi-drug resistant infections. Ame Res J Biotechnol. 2017c;1(1):1-9.
- Chakraborty AK. Mechanism of AMR: MDR genes and antibiotics decoys retard the new antibiotic discovery against superbugs Novel Appo Drug Des and Dev. 2017d;2(1): 555576.
- Chakraborty AK. Multi-drug resistant bacteria from Kolkata Ganga river with heterogeneous MDR genes have four hallmarks of cancer cells but could be controlled by organic phyto-extracts. Biochem Biotechnol Res.2017e;5(1):11-23.
- Chakraborty AK. High mode contamination of multi-drug resistant bacteria in Kolkata: Mechanism of gene activation and remedy by heterogeneous phyto-antibiotics. Ind J Biotechnol. 2015;14:149-159.
- Huang TW, Wang JT, Lauderdale TL et al. Complete sequences of two plasmids in a blaNDM-1-positive Klebsiella oxytoca isolate from Taiwan. Antimicrob Agents Chemother. 2013;57(8):4072-76.
- Chakraborty AK. MDR Genes are created and transmitted in plasmids and chromosomes to keep normal intestinal microbiota alive against high dose antibiotics- A Hypothesis. J Mol Med Clin Appl. 2017a;2(1).
- Johnson M, Zaretskaya I, Raytselis Y, et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36 (suppl 2):W5-W9.
- Hu H, Hu Y, Pan Y, et al. Novel plasmid and its variant harboring both a blaNDM-1 Gene and Type IV Secretion System in clinical Isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother. 2012;56(4):1698-1702.
- Khan AU, Rehman MT. Role of Non-Active-Site Residue Trp-93 in the Function and Stability of New Delhi Metallo-β-Lactamase 1. Antimicrob Agents Chemother. 2015;60(1):356-60.
- Manageiro V, Sampaio DA, Pereira P, et al. Draft genome sequence of the first NDM-1 producing Providencia stuartii strain isolated in Portugal. Genome Announc. 2015;3(5): e01077-15.
- Mataseje LF, Peirano G, Church DL, et al. Colistin non-susceptible Pseudomonas aeruginosa ST654 with blaNDM-1 arrives in North America. Antimicrob Agents Chemother. 2016;AAC.02591-15.