Genetic characterization of s1 domain of porcine epidemic diarrhea viruses spike proteins isolated in Korea
2 Department of Veterinary Microbiology and Infectious Diseases, Faculty of Veterinary Medicine, Vietnam National University of Agriculture, Hanoi, Viet Nam, Email: giapnv@ueh.edu.vn
3 Research Unit, Green Cross Veterinary Products, Yongin, Republic of Korea, Email: mhj1219@naver.com
4 Viral Infectious Disease Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon 305-806, Republic of Korea, Email: kimkwonhye@yahoo.com
5 Forensic Medicine Division, Daegu Institute, National Forensic Service, Chilgok, 718-803, Republic of Korea, Email: parkjunseong@gmail.com
Bong Kyun Park, Department of Veterinary Medicine Virology Lab, College of Veterinary Medicine and Research Institute for Veterinary Science, Seoul National University, DaeHakRo, GwanAk-Gu, Seoul 151-742, Korea, Tel: +8228801255, Fax: +8228850263, Email: parkx026@snu.ac.kr
Received: 22-Aug-2017 Accepted Date: Aug 31, 2017; Published: 06-Sep-2017
Citation: Chung HC, Nguyen VG, Moon HJ, et al. Genetic characterization of s1 domain of porcine epidemic diarrhea viruses spike proteins isolated in korea. J Immune Disord Ther 2017;1(1)1-3.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
We analyzed spike gene of the three PEDV isolated during 2014 to 2015 in our lab (Strain J3142, BM1, and BM3). In phylogenetic study, 3 strains belong to gene subgroup G2a, and between three strains, BM1 and BM3 have higher similarity with the strains from North America. On the other hand, J3142 was more closely related to China strain and has some variation in the S1 region (1,938- 2,077nt) compared to BM strains. Especially from the data, analyzing N-terminal domain and C-terminal domain, N-terminal domain showed little homology to strains from USA and China.
Keywords
S1 domain; Porcine Epidemic Diarrhea Virus; South Korea; Isolation
Porcine epidemic diarrhea (PED) is an acute contagious diarrhea disease caused by porcine epidemic diarrhea virus (PEDV) in pigs [1]. Since PEDV was first reported in South Korea in 1992 [2], it has been circulating so far, and exhibited significant genetic diversity [3]. For 2 decades, PEDV was reported in several European countries; Hungary, France, Canada, Italy, and Switzerland [1].
The genome of PEDV consists of a single stranded RNA with positive polarity. It is approximately 30,000 nucleotides in length and contains a cap structure at the 51 end and a poly (A) tail at the 31 end. In the order from 51 to 31, the genome of PEDV contains the genes for the spike (S) protein, an open reading frame (ORF3), the small membrane protein (sM), M protein, and nucleocapsid (N) protein [4].
Especially, glycoprotein of S gene on the viral surface plays a significant role in induction of neutralizing antibodies which bind to specific receptors and induce cell membrane fusion [5-7]. It was divided into two domains, S1 and S2; the former is responsible for binding to the host–specific receptor, while the latter appears to be involved in the direct fusion process between viral and cellular membranes [7,8]. Although vaccines (DR13, SM98 strains) have been developed and used in commercial swine farms for the prevention of the disease, it has been reported that recently isolated strains in South Korea and those in the United States had S1 gene variant region and were closely related to each other; therefore, the efficacy of vaccines is now doubtful [9,10]. Recently, it became a more important pathogen because the virus occurred endemics in Asia and became more acute and severe form [1,10]. Besides, it has severely attacked the United States in April 2013, and led to significant economic losses in the pig industry even the U.S.A. has adopted the way of import restriction [11-13]. These strains detected in the United States showed significant similarity to a China strain, AH2012, showing 99.5% identity [14,15]. Until now, Ohio and new US strains were reported to show genetic diversity causing clinical symptoms [16]. Based on seroprevalence from the samples collected in 2012-2013, most of PEDV in South Korea showed infection patterns in sucling and weaned piglets similar with those of PEDV in North America. However, it became infect in all ages of pigs and highly increasing events after the report of outbreak in North Korea after 2014 [17].
In this study, we will report some field the isolates (J3142, BM1 and BM3) of the emerging PEDVs and their genetic relationships with other strains.
Between 56 PED positive samples, three (J3142; KP995064, BM1; KP861982, and BM3; KP995065) strains were used for this research, which were isolated among 56 PED positive samples. 411 reference sequences (Supplement Table S1) including S gene of PEDV strains were collected from the GenBank database which contain currently detected strains in the United States and Asia. Acquired nucleotide sequences in this study and reference sequences were aligned using the CLUSTAL W. The construction of phylogenetic tree was performed using a maximum likelihood with bootstrap replicates 1000 by Fasttree [18]. Also, it was conducted to analysis both N-terminal domain (NTD) (1-494aa) and C-terminal domain (CTD) (495–697 aa) of the S1 gene region [19] which is playing as important binding receptor with above references sequences. Percentage identities of the aligned complete and NTD of S spike protein were shown in the upper and lower triangles, respectively.
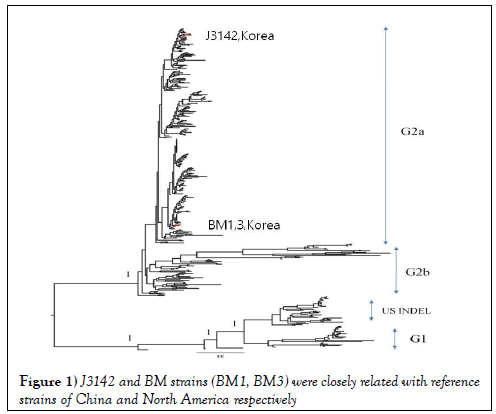
In phylogenetic analysis, S gene complete sequences (BM1, BM3) detected in this study was put together with PEDVs in the United States, which belongs to subgroup 2a, genogroup 2 (99.5~99.7% similarity). Although J3142 strain is in the same branch of genogroup 2, it revealed that J3142 was more closely related to China root strains (AH2012, BJ2012-2) showing 98.5~98.9% identity than strains of the United States (Figure 1, Supplement Figure S1).
From genetic and phylogenetic analyses of S gene of newly isolates, J3142 and BM strains (BM1, BM3) were closely related with reference strains of China and North America respectively, however, both were located in a different genogroup with currently used vaccine strains (CV777 and DR13).
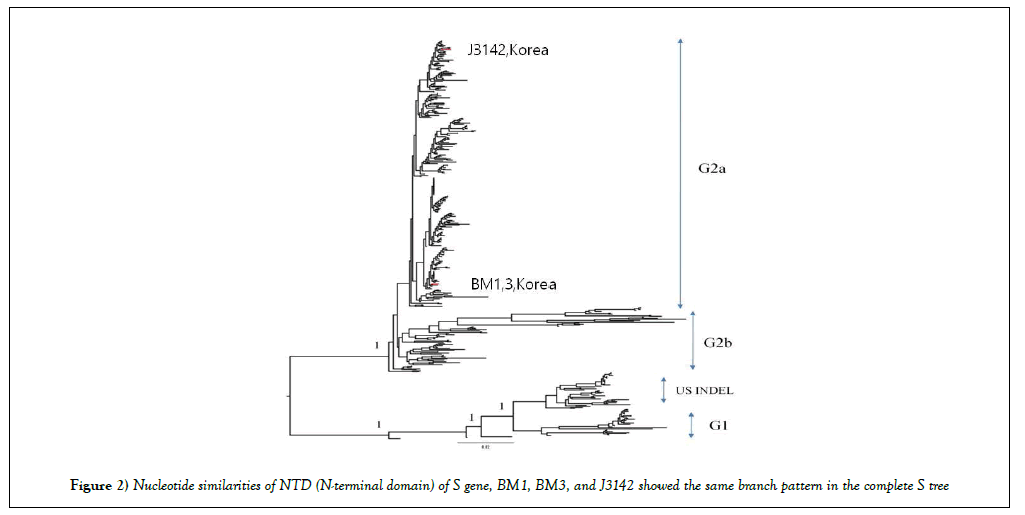
Next, in nucleotide similarities of NTD (N-terminal domain) of S gene, BM1, BM3, and J3142 showed the same branch pattern in the complete S tree. It was suggested that the only N-terminal region of S1 domain would be sufficient for revealing genetic relatedness among different PEDV isolates [11,20]. On the other hand, they had lower similarities with DR13 and CV777 (genogroup 1 derived), vaccine strains in Korea, showed 89.5% and 89.2% identities respectively (Figure 2 and Table 1). Between complete and NTD complete S gene sequences, NTD region was more different than complete S gene region (Figure 2 and Supplement Figure S2).
 |
||||||||||
| DR13 | BM1 | J3142 | BM3 | CV777 | AH2012 | BJ-2012-2 | Colorado30 | Indiana34 | Texas31 | |
|---|---|---|---|---|---|---|---|---|---|---|
| DR13 | 94.5 | 93.9 | 94.5 | 98.3 | 94.7 | 94.8 | 94.6 | 94.6 | 94.7 | |
| BM1 | 89.4 | 98.2 | 99.9 | 93.4 | 99.0 | 99.2 | 99.7 | 99.5 | 99.7 | |
| J3142 | 89.5 | 98.4 | 98.2 | 92.8 | 98.5 | 98.9 | 98.3 | 98.1 | 98.4 | |
| BM3 | 89.4 | 100 | 98.4 | 93.3 | 99.0 | 99.2 | 99.7 | 99.4 | 99.7 | |
| CV777 | 98.2 | 89.2 | 89.3 | 89.2 | 93.6 | 93.7 | 93.5 | 93.4 | 93.5 | |
| AH2012 | 89.7 | 98.8 | 99.1 | 98.8 | 89.5 | 99.5 | 99.1 | 98.8 | 99.2 | |
| BJ-2012-2 | 89.8 | 98.9 | 99.0 | 98.9 | 89.6 | 99.2 | 99.3 | 99.1 | 99.3 | |
| Colorado30 | 89.4 | 99.7 | 98.4 | 99.7 | 89.2 | 98.8 | 98.9 | 99.6 | 99.8 | |
| Indiana34 | 89.6 | 99.1 | 98.0 | 99.1 | 89.3 | 98.3 | 98.4 | 99.1 | 99.6 | |
| Texas31 | 89.6 | 99.7 | 98.6 | 99.7 | 89.4 | 99.0 | 99.0 | 99.7 | 99.2 | |
Table 1: Nucleotide both Complete and N-terminal domain (NTD) of the S gene between strains in the Korea, China, and United States. Percentage identities that complete and NTD of S gene the aligned nucleotide were shown in the upper and lower triangles, respectively
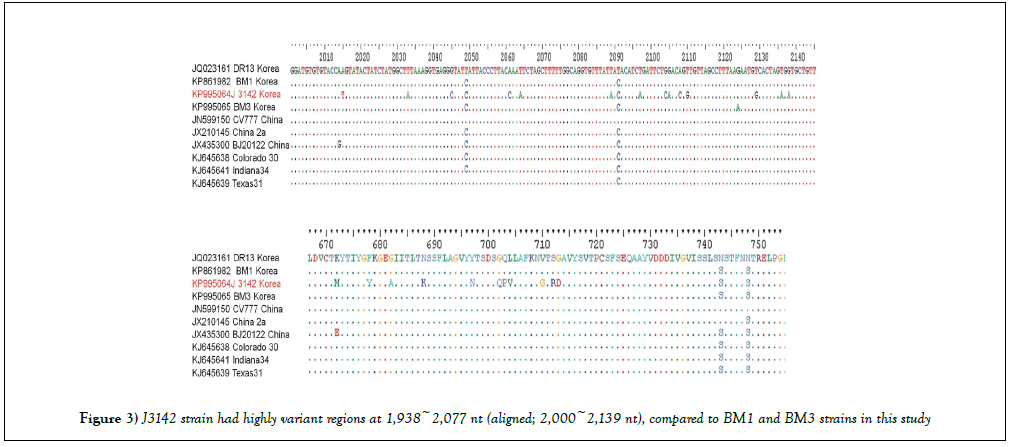
CTD region showed homology of 95.5-97.1% with current circulating strains of South Korea, China, and United States (Supplementary Table S2). Especially, J3142 strain had highly variant regions at 1,938~2,077 nt (aligned; 2,000~2,139 nt), compared to BM1 and BM3 strains in this study (Figure 3).
The S1 N-terminal region of PEDVs circulating in Korea was used to construct the phylogenetic tree (Figure 2 and Table 1). It was suggested that the only N-terminal region of the S1 domain would be sufficient for revealing genetic relatedness among different PEDV isolates [11,12,15].
The attenuated PEDV vaccines based on the CV777 or DR13-derived strains might be antigenically less related to the newly emergent PEDV strains; thus, new vaccines based on current strains are needed [8,21].
In the tree, PEDV isolates in South Korea were highly similar to those detected in the USA. This situation similarly occurred in the USA, June, 2013 [12]. Although there was an import restriction on pig or pork products from other countries with endemic or epidemic PED, it has shown that the strains were closely related to the strain of China, AH2012 (KC210145) [22-24]. It may suggest that epidemiological and retrospective studies are necessary to identify the source of PED in South Korea as well as in the USA.
Until now, many strains of the Unite States, China, and South Korea were reported as S gene variant strains. It shows that molecular based analysis is important to prevent severe economic loss in swine farms.
In conclusion, after the PED outbreaks in the United States, PED have also explosively occurred in South Korea. The current strains in Korea would be highly similar to those in the United States, throwing the question about epidemiological sources. Although strains of Korea and the United States were genetically closely related to each other, prevalence and virulence pattern were different, which suggests that other factors such as the environment or vaccination would be associated. Therefore, additional epidemiological studies should be performed with molecular S gene genetic analysis.
Acknowledgements
The authors would like to thank Hye Jung Yang and Jung Ah Kim for excellent technical assistance. This study was supported by the Bio Green 21 Program, Rural Development Administration (Grant no. PJ011184), and by the Bioindustry Technology Development Program (grant no. 114055031SB010), Ministry of Agriculture, Food and Rural Affairs, South Korea.
Conflict of Interest
The authors declare that there are no conflicts of interest.
REFERENCES
- Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44(2):167-75.
- Kweon CH, Kwon BJ, Jung TS, et al. Isolation of porcine epidemic diarrhea virus (PEDV) in Korea. Korean J Vet Res 1993;33(2):249-54.
- Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes 2012;44(2):167-75.
- Kweon CH, Kwon BJ, Jung TS, et al. Isolation of porcine epidemic diarrhea virus (PEDV) in Korea. Korean J Vet Res 1993;33(2):249-54.
- Choi JC, Lee KK, Pi JH, et al. Comparative genome analysis and molecular epidemiology of the reemerging porcine epidemic diarrhea virus strains isolated in Korea. Infection, Genetics and Evolution 2014;26:348-51.
- Tobler K, Bridgen A, Ackermann M. Sequence analysis of the nucleocapsid protein gene of porcine epidemic diarrhoea virus. In Coronaviruses. Springer 1994; pp: 49-54.
- Cruz DJ, Kim CJ, Shin HJ. Phage-displayed peptides having antigenic similarities with porcine epidemic diarrhea virus (PEDV) neutralizing epitopes. Virology 2006;354(1):28-34.
- Duarte M, Laude H. Sequence of the Spike Protein of the Porcine Epidemic Diarrhoea Virus. Journal of General Virology 1994;75(5):1195-200.
- Lee DK, Cha SY, Lee C. The N-terminal region of the porcine epidemic diarrhea virus spike protein is important for the receptor binding. Korean J Microbiol Biotechnol. 2011;39(2):140-5.
- Gallagher TM, Buchmeier MJ. Coronavirus spike proteins in viral entry and pathogenesis. Virology 2001;279(2):371-4.
- Chung HC, Nguyen HJ, Lee JH, et al. Isolation of porcine epidemic diarrhea virus during outbreaks in South Korea, 2013–2014. Emerging Infectious Diseases 2015;21(12):2238.
- Tian Y, Yu Z, Cheng K, et al. Molecular characterization and phylogenetic analysis of new variants of the porcine epidemic diarrhea virus in Gansu, China in 2012. Viruses. 2013;5(8):1991-2004.
- Chen Q, Li G, Stasko J, et al. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. Journal of Clinical Microbiology 2014;52(1):234-43.
- Cima G. Viral disease affects US pigs: porcine epidemic diarrhea found in at least 11 states.
- Mole B. Deadly pig virus slips through US borders. Nature 2013;499(7459):388.
- Huang YW, Dickerman AW, Piñeyro P, et al. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio 2013;4(5):e00737-13.
- Jung K, Wang Q, Scheuer KA, et al. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerging Infectious Diseases 2014;20(4):662.
- Oka T, Saif LJ, Marthaler D, et al. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Veterinary microbiology. 2014;173(3):258-69.
- Park B, Song D. Recent Outbreaks and emergence of mutants of Porcine Epidemic Diarrhea Viruses (PEDV) in Korea. Japanese Journal of Veterinary Research. 2016;64(Supplement 1):S25-32.
- Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PloS ONE 2010;5(3):e9490.
- Lee DK, Park CK, Kim SH, et al. Heterogeneity in spike protein genes of porcine epidemic diarrhea viruses isolated in Korea. Virus Research 2010;149(2):175-82.
- Vlasova AN, Marthaler D, Wang Q, et al. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerging Infectious Diseases 2014;20(10):1620.
- Wang L, Byrum B, Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerging Infectious Diseases 2014;20(5):917.
- Vlasova AN, Marthaler D, Wang Q, et al. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerging Infectious Diseases 2014;20(10):1620.