Marine microorganisms: Evolution and solution to pollution
2 University of Chinese Academy of Sciences, Beijing, 100039, China
Received: 26-Dec-2017 Accepted Date: Jan 08, 2018; Published: 18-Jan-2018
Citation: Li FL, Wang B. Marine microorganisms: Evolution and solution to pollution. J Mar Microbiol. 2018;2(1):4-5.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Once ocean nurtured life, now she needs our care. Marine microorganism is the host of ocean in all ages. We should learn from them humbly. Marine microorganism is tightly bond with human during the history of evolution and nowadays’ environment pollution.
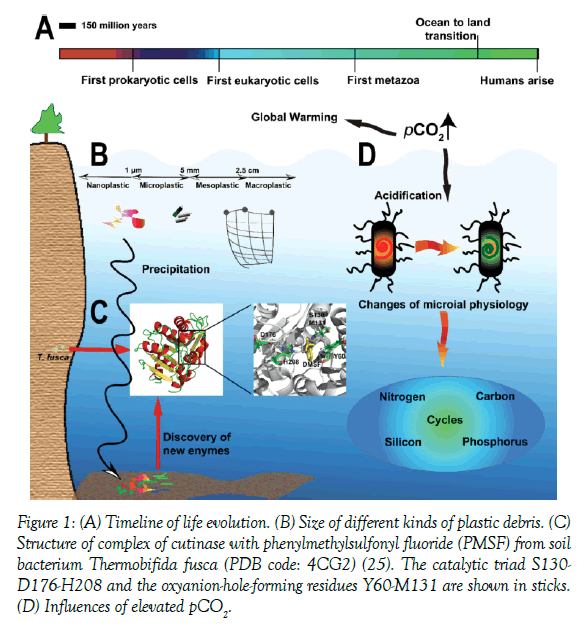
Although the topic is still in debate, life is probably originated from submarine in hydrothermal vent systems [1]. In the journey of evolution, our biosphere was completely dominated by microbes for a very long time (Figure 1A). Human being evolves with those microorganisms. Consequently, the influences of microorganisms can be found in all aspects of human biology. More than 65% of our genes originated with bacteria, archaea, and unicellular eukaryotes, including those genes responsible for host-microbe interactions. Recently, the microbiota has been regarded as a new organ of human being [2]. Cross-talk between us and gut microbiota affects both our physical and psychological health [3,4].
Figure 1: (A) Timeline of life evolution. (B) Size of different kinds of plastic debris. (C) Structure of complex of cutinase with phenylmethylsulfonyl fluoride (PMSF) from soil bacterium Thermobifida fusca (PDB code: 4CG2) (25). The catalytic triad S130- D176-H208 and the oxyanion-hole-forming residues Y60-M131 are shown in sticks. (D) Influences of elevated pCO2.
Although evolution makes us to be immune to many pathogens, till now humans and animals still suffer from many old infections. For example, Helicobacter pylori have coevolved with us for at least 50,000 years [5,6]. Studying the physiological-biochemical properties of ancient marine bacteria will be an opportunity to further understand ourselves and to seek for new methods of fighting old infections.
Along with industrial revolution, our marine ecosystem suffered serious pollutions. Microplastics are tiny plastic particles (<5 mm) (Figure 1B), which poison marine lives. Because these microplastics are very hard to be degraded, it is predicted that there will be more microplastics than fish in ocean by the year 2050 [7]. Since marine sediments are considered as the sink of microplastics and marine microbes are key dwellers of marine sediments, more attention should be paid on the interactions between microplastics and marine microbes. Actually, a call for this has been published in 2011 [8]. Searching for marine bacteria which can digest microplastics is one of the solutions to clean marine environments. For example, a polyethylene terephthalate (PET) utilization bacterium Ideonella sakaiensis was isolated. It can adhere to PET and secretes PET hydrolase (PETase) to target ester bonds. Then mono-(2-hydroxyethyl) terephthalic acid (MHET) is produced, which will be rapidly hydrolyzed by MHET hydrolase to monomers, terephthalic acid and ethylene glycol [9]. Recently, the structure of a PETase from I. sakaiensis was solved, and a catalytic mechanism was proposed [10]. Getting insight into the catalytic mechanism of plastic-degrading enzymes helps us to engineer novel enzymes (Figure 1C). Except bacteria, many fungi which can degrade polyethylene microplastics were also isolated and characterized [11,12]. Study dynamics of microbial communities is essential. A study found that the biofilm formation of a bacterial and fungal community on plastic debris could be hampered in natural environment [13]. As knowledge increases, we could build a microbial platform for biological recycling of PET.
Marine macroalgae, fixing abundant CO2, is a promising feedstock for biofuel production. Our lab isolated a novel thermophilic bacterium Defluviitalea phaphyphila Alg1 from coastal sediments, and the brown alga-degradation systems of Alg1 was analyzed [14]. Alg1 is able to directly ferment 5% unpretreated seaweed powder to produce 10 g/L ethanol at 60°C [15]. Alginate is one of the three main sugars of brown algae (the other two are mannitol and laminarin), which can be depolymerized by polysaccharides lyases via β-elimination reaction. Although many alginate lyases have been characterized, a few of them are thermophilic [16]. A thermophilic polysaccharide lyase family 6 alginate lyase from Alg1 was very intriguing. And we found that one substitution of aspartate for glutamate in the calcium-binding sites led to enhanced substrate affinity [17], which might be a result of thermophilic adaption.
The increased partial pressure of CO2 (pCO2) leads to global warming not only, but also the acidification of ocean. By the end of 21st century, H+ concentration of surface oceans will be as twice as those of the preindustrial ocean. How acidification of ocean affects the evolution of marine microbes is still lack of solid evidences [18,19]. For now, only a few biomineralizing microbes have deep-time studies of evolutionary responses to global climate changes. However, this acidification has complex effects on ocean cycles of carbon, nitrogen, phosphorus, and silicon through changing the microbial physiology (Figure 1D) [20]. For example, nitrogen fixation and growth of cyanobacterium Trichodesmium spp. decreased under acidified environment, though they could be enhanced by high pCO2 [21,22]. Although phytoplankton was likely to accumulate more biomass, they would undergo iron stress caused by ocean acidification [23]. Thus the influences of ocean acidification are very uncertain. In the future, we should keep observing the effects of ocean acidification on marine life. Meanwhile, because algae can sever as CO2 sink, developing algae industry will be one way to decrease pCO2.
Microbiome is a rising star today. Many related projects have been launched, for example, the Earth Microbiome Project [24,25]. Using this powerful tool, we can rapidly analyze the structure of marine microbial ecosystems and the influence of human activities on these ecosystems. Besides, the metagenome data will speed up the isolation of novel microbes and genes, which help us to deeply understand and friendly live with the ocean.
REFERENCES
- Bassez MP. Is high-pressure water the cradle of life? J Physics: Condensed Matter. 2003;15(24):353.
- Rook G, Bäckhed F, Levin BR, et al. Evolution, human-microbe interactions, and life history plasticity. The Lancet. 2017;390(10093):521-30.
- Foster JA, Neufeld KAM. Gut–brain axis: how the microbiome influences anxiety and depression. Trends in Neurosciences. 2013;36(5):305-12.
- Parker A, Lawson MA, Vaux L, et al. Host‐microbe interaction in the gastrointestinal tract. Environmental Microbiology. 2017.
- Atherton JC, Blaser MJ. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. The J Clinical Investigation. 2009;119(9):2475.
- Achtman M. How old are bacterial pathogens? Proceedings Biological Sci. 2016;283(1836):20160990.
- Auta H, Emenike C, Fauziah S. Distribution and importance of microplastics in the marine environment: A review of the sources, fate, effects, and potential solutions. Environment International. 2017.
- Harrison JP, Sapp M, Schratzberger M, et al. Interactions between microorganisms and marine microplastics: a call for research. Marine Technology Society J. 2011;45(2):12-20.
- Yoshida S, Hiraga K, Takehana T, et al. A bacterium that degrades and assimilates poly (ethylene terephthalate). Science. 2016;351(6278):1196-99.
- Han X, Liu W, Huang JW, et al. Structural insight into catalytic mechanism of PET hydrolase. Nature Communications. 2017;8(1):2106.
- Paço A, Duarte K, da Costa JP, et al. Biodegradation of polyethylene microplastics by the marine fungus Zalerion maritimum. Science of The Total Environment. 2017;586:10-15.
- Pramila R, Ramesh KV. Biodegradation of low density polyethylene (LDPE) by fungi isolated from municipal landfill area. J Microbio Biotech Res. 2017;1(4):131-6.
- De Tender CA, Devriese LI, Haegeman A, et al. The temporal dynamics of bacterial and fungal colonization on plastic debris in the North Sea. Environmental Science & Technology. 2017.
- Ji SQ, Wang B, Lu M, et al. Defluviitalea phaphyphila sp. nov., a novel thermophilic bacterium that degrades brown algae. Applied and Environmental Microbiology. 2016;82(3):868-77.
- Ji SQ, Wang B, Lu M, et al. Direct bioconversion of brown algae into ethanol by thermophilic bacterium Defluviitalea phaphyphila. Biotechnology for Biofuels. 2016;9(1):81.
- Wang B, Ji SQ, Lu M, et al. Biochemical and structural characterization of alginate lyases: an update. Current Biotechnology. 2015;4(3):223-39.
- Wang B, Ji SQ, Ma XQ, et al. Substitution of one calcium-binding amino acid strengthens substrate binding in a thermophilic alginate lyase. FEBS Letters. 2018.
- Hutchins DA, Fu F. Microorganisms and ocean global change. Nature Microbiology. 2017;2:17058.
- Sunday JM, Fabricius KE, Kroeker KJ, et al. Ocean acidification can mediate biodiversity shifts by changing biogenic habitat. Nature Climate Change. 2017;7(1):81.
- Hutchins DA, Mulholland MR, Fu F. Nutrient cycles and marine microbes in a CO2-enriched ocean. Oceanography. 2009;22(4):128-45.
- Levitan O, Rosenberg G, Setlik I, et al. Elevated CO2 enhances nitrogen fixation and growth in the marine cyanobacterium Trichodesmium. Global Change Biology. 2007;13(2):531-8.
- Hong H, Shen R, Zhang F, et al. The complex effects of ocean acidification on the prominent N2-fixing cyanobacterium Trichodesmium. Science. 2017;356(6337):527-31.
- Shi D, Xu Y, Hopkinson BM, et al. Effect of ocean acidification on iron availability to marine phytoplankton. Science. 2010;327(5966):676-9.
- Gilbert JA, Jansson JK, Knight R. The Earth Microbiome project: successes and aspirations. BMC Biology. 2014;12(1):69.
- Roth C, Wei R, Oeser T, et al. Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Applied Microbiology and Biotechnology. 2014;98(18):7815-23.