Non Random distribution of Chromosome -21 in Syndromic Cases of Wilms’ tumors
2 Department of Paediatric Surgery, All India Institute of Medical Sciences, Bihar, India
3 Department of Radiotherapy, All India Institute of Medical Sciences, Bihar, India
Received: 08-Jun-2020, Manuscript No. PULJCGG-20-2682; Editor assigned: 10-Jun-2020, Pre QC No. PULJCGG-20-2682(PQ); Reviewed: 24-Jun-2020 QC No. PULJCGG-20-2682; Revised: 18-Nov-2022, Manuscript No. PULJCGG-20-2682(R); Published: 16-Dec-2022
Citation: Saxena AK, Kumar V, Singh P, et al. Non random distribution of chromosome-21 in syndromic cases of Wilms’ tumors. J Clin Gen Genomics 2022;5(1):1-4.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Wilms’ tumors are the common embryonic childhood tumor and heterogeneous in nature. Karyotypic analysis of Wilms’ tumor has shown a variety of chromosomal aberrations with different (%) frequency of structural and numerical changes in somatic cell. Present study were carried out in eleven cases (n=11) of Wilms’ tumors with different age group (betwen 1.5 year to 10 year). Most striking feature is the involvement of high frequency (>60%) of Wilms’ tumor shows extra copy of chromosome -21 in the karyotypes after using high resolution of GTG banding and FISH analysis. Interestingly, 18% cases of Wilms’ tumor shows loss of Ychromosome and appearance of (r) Y reporting first time in India. Beside this, short arm of chromosome-6 and 16 shows two new break points i.e. 6q21.22 and 16q23 might have play an significant role in Wilms’ tumors progression. The another relevant were the association of either gain (trisomy) or loss (monosomy) of chromosome with chromatid break points, ring, dicentric or rearrangements of chromosome (translocation) with different frequency. However, this is a rare coincidence that environmental factor (s) might have increase risk of developing down syndrome (47, XY+21) in Wilms’ tumors, due non-disjunction event and unequal crossing over in cell- division of the disease outcome.
Keywords
Wilms’ tumor; Trisomy -21; Heterozygocity; Down syndrome
Introduction
Wilms’ Tumor (WT) is common embryonic tumour of paediatric age group (1-5 year) [1]. The incidence of Wilms’ tumor is 1 per 10,000 and more than 90% cases are non familial (sporadic) and 1%-2% are familial (heritable) in nature [2]. Several studies exist on cytogenetics basis with genetic diversity in term of chromosomal aberrations and loss of heterozygocity in Wilms’ tumour [3]. Variety of Chromosome Aberrations (CAs) were observed including short arm of chromosome-11p deletion at two different regions i.e. 11p13 and 11p15 besides involvement of 1p, 4q, 7p, 11q, 14q, 16q and 17p [4]. Beside these numerical variations i.e. monosomies of chromosome-16, 22, X and trisomy of chromosome - 6,7,8,13,13, 18 were observed in different karyotypes of Wilms’ tumors [5].
The pathogenesis of Wilms’ tumors appears is highly complex due to involvement of several genes loci distributed at different chromosomes [6].
The concept of deletion of WT1 gene locus at chromosome 11p13 with Loss of Heterozygocity (LOH) is frequently involved in the children of Wilms’ tumor with WAGR syndrome (Wilms-Aniridia-Genitourinary anomalies-Mental retardation) and only 20% cases are non-syndromic. Classically, WTI gene is also known as tumour suppressor gene and first mapped on chromosome 11p13 and has been mutated in more than 10% sporadic cases of Wilms’ tumours [7]. It is still not clear that how constitutional mutation of WT1 gene have been associated to either unilateral or bilateral disease with congenital genitourinary disorders in the family of Wilms’ tumour [8]. The other constitutional genetic changes occurs due to loss of WTI gene can predispose in rare syndromes (less than 1%) such as Li-Fraumeni (TP53), Fanconi anaemia (BRCA2), CLOVES (PIK3CA) and Perlman (DIS3L2) [9]. Several other loci of different chromosomes -1p, 2q, 7p, 9q, 14q, 11p15, 16q, 22q suggest the Loss Of Heterozygocity (LOH) and existence of tumour suppressor gene have been known and associated with different etiopathology of Wilms’ tumour predisposition [10]. However, such accumulating evidence suggests that the progression of various tumors to complete malignancy requires genetic alterations which activate cellular oncogenes or inactivate tumor suppressor genes or mismatch DNA repair mechanism [11]. The loss of genetic material at distal region of chromosome-7p22 leads to somatic mutation and develop constrains of putative tumor suppressor gene(s) [12]. Trisomy of chromosomes- 6, 8 and 12 being the most frequent numerical and structural variation of 1p, 7p, 11p and 16q break points (sites) have also been associated in progression of Wilms’ tumors. There is scarcity in the literature that Wilms’ tumor haven associated to down syndrome and turnor syndrome [13].
In the present study we describe the spectrum of complex non- random involvement of chromosome abnormalities with high frequency of chromosome-21 karyotypes was predominant besides appearance of ring of Y and loss of Y-chromosome in karyotypes, reporting first time in the cases of Wilms’ tumors in eastern part of India [14]. Hence, the study becomes relevant to discuss, the role of these genetics variations during development and progression of Down Syndrome (DS) in Wilms tumors [15].
Case Presentation
Patients
Our study included clinically diagnosed cases (n=11) of Wilms Tumor (WT) referred to the cytogenetic and molecular laboratory, Department of pathology/lab medicine, All india institute of medical sciences patna for genetic analysis. Family history was recorded to develop pedigree analysis and to find out the mode of inheritance in the proband [16]. Family history showing lack of environmental exposure either to radiation or prenatal exposure to drug involvement during tumor development [17]. Median age of the proband was 5.7 years (range 1.5-10 years). Blood samples (1.0 ml) were collected under sterile conditions in heparinised vial. The patients gave informed written consent for cytogenetic analysis. The present study was approved by the Institutional Ethics Committee (IEC) of the Institute [18].
Karyotyping and fluorescence in situ hybridization analysis
Karyotypes was developed from proband using short term lymphocytes cultures with RPMI 1640 media containing phytohaemagglutinin, (5%) FBS and 1% antibiotics solution (streptomycin-penicillin) for 72 hours at 37°C.
Before harvesting the cultures, colchicine was added 2 hrs prior to arrest the dividing cells [19]. Prewarmed KCl solution was used as hypotonic and cells were fixed in 3:1 methanol: Acetic acid solution. At least twenty well spread metaphases were selected for karyotypes after GTG banding [20]. The karyotypes were prepared according to the recommendations of the International System for Chromosome Nomenclature (ISCN 2016) using applied spectral imaging software (Genesis USA) [21]. FISH analysis was carried out for further confirmation of extra copy of chromosome-21 in both interphase as well as metaphases using FISH DNA probe labeled with Spectrum Orange of 220 kb and cytogenetic location 21q22.13-q22.21 obtained from Abbott-Vysis, Inc. (USA). All details concerning hybridization were carried out according to the instructions provided in the kit. The chromosome were counterstained with DAPI and viewed under fluorescence microscope (Olympus Japan). More than 100 interphase cells and 5-10 metaphase were recorded for analysis [22].
Statistical analysis
X2-test (Chi square) were used to find out level of significance difference (pvalue) between the normal and abnormal karyotypes in the cases of Wilms tumor [23].
Results
Clinically diagnosed cases (n=11) of Wilms’ tumor and out of which nine were (n=9) males and (n=2) were females with mean age group 5.7 years included in the present study to evaluate the spectrum of chromosomal aberrations and their (%) frequency using short term lymphocyte culture are documented in Table 1 [24].
| S.No | Age /Gender | Karyotyping variations | Total (%) frequency of abnormal cells |
|---|---|---|---|
| 1 | 5/M | 46,XY | 0 |
| 2 | 1.5/M | 45,X0 | 60 |
| 3 | 5Y/M | 47,XY,+21,Y-ring/45,XY,-22/47,XY,+9/45,XY,-18 | 66.6 |
| 4 | 10/M | 46,XY ,brk 16q23 ,brk 6q21.22 /47,XY,+21 /45,XY,-22/45,XY,-16 | 60 |
| 5 | 7/M | 47,XY,+18/47,XY,+11/47,XY,+9 /45,XY,-16 /47,XY,+18 /45,XY-11 /46,XY,dicen 15 | 38 |
| 6 | 9/M | 47,XY,+17/47,XY,+21 | 50 |
| 7 | 3/M | 45,XY,-22/45,XY,-19 /46,XY,Y-ring /45,XY,-15/45,XY,-18/45,XY,-11 /47,XY,+17/47,XY,+21 | 53 |
| 8 | 4.5/M | 47,XY,+21 | 33.33 |
| 9 | 10/F | 45,XX,reci.trans.(15-21) | 20 |
| 10 | 4/F | 47,XX,+21 | 70 |
| 11 | 8/M | 47,XY,+21 /49,XY,+18,+21,+22 | 44.44 |
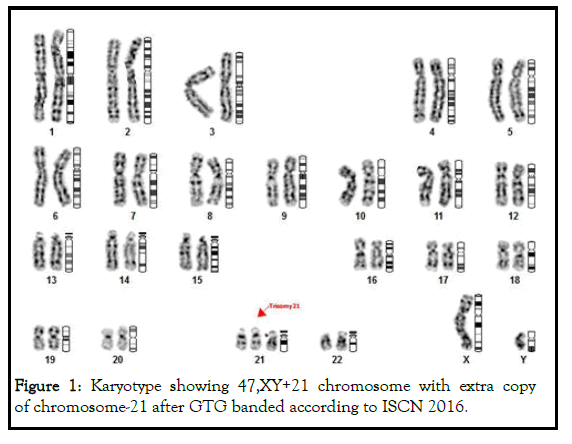
More than sixty (>60%) cases of Wilms tumors shows extra copy of chromosome-21 in the karyotypes (47,XY+21) and designated as down syndrome (Figure 1).
Figure 1: Karyotype showing 47,XY+21 chromosome with extra copy of chromosome-21 after GTG banded according to ISCN 2016.
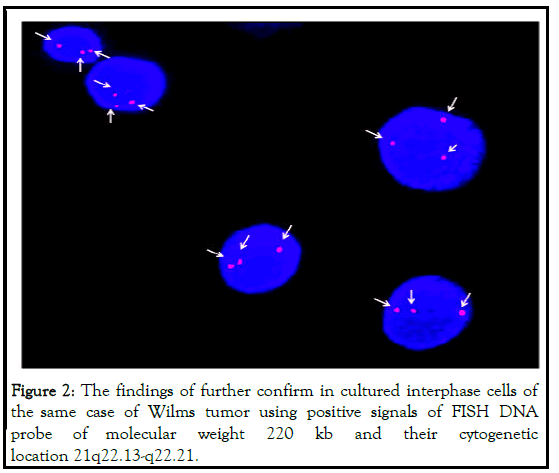
It was further confirmed by Fluorescence in situ Hybridization (FISH) using specific probe as visualized in Figure 2 [25].
labeled with spectrum orange (red) and DAPI (blue) is used as nuclear stain for back ground staining as shown in Figure 2.
In cultured interphase cells (arrow head). One cases of Wilms tumor (~9%) showing (47,XY+18) karyotype diagnosed as Edward syndrome [26].
Interestingly, these cases showing lack of phenotypic appearance of syndrome or any preclinical sign of congenital anomaly [27]. The study of non- random inter and intra chromosomal variations in Wilms tumor becomes more relevant due to increase incidence (6%) of mosaicism having 46, XY/47,XY or 46XY/45,XY in karyotypic pattern between syndromic (>7%) or non- syndromic (>2%) cases of Wilms tumors [28]. The karyotypic variation (s) in individual cases of Wilms tumor is described below:
Case 1: A five year male having both structural and numerical chromosomal variations in the karyotypes- 47,XY+21; 47,XY,+9; 45,XY,-18; 45,XY,-22; 47,XY+21+(r)Y.
Case 2: A ten year male showing structural and numerical chromosomal abnormalities in the karyotypes- 47,XY+21; 46,XY,cht brk 16q23; 46,XY,ch brk 6q21.2; 45,XY-16;45,XY-22; 46,XY, aneuploidy.
Case 3: A seven year male having structural and numerical chromosomal abnormalities in karyotypes-47,XY,+9; 47,XY,+11; 47,XY,+18 (3);45,XY-11; 45,XY-16; 46,XY+dic(15).
Case 4: A three year male showing both structural and numerical chromosomal abnormalities in karyotypes - 47,XY+21; 45,XY,-11; 45,XY,-15; 45,XY,-17; 45,XY,-18; 45,XY,-19; 45, XY,-22; 46,X(r)Y.
Case 5: A nine year male showing numerical chromosomal abnormalities in the karyotypes-47,XY+17; 47,XY+ 21 (2).
Case 6: A four year and six month old male showing numerical chromosomal abnormality in karyotypes-47,XY+21 (Trisomy-21).
Case 7: A eight year male showing structural and numerical chromosomal abnormalities in karyotypes-47,XY+21 (4); 49,XY+18,22.
Case 8: A ten year male showing numerical chromosomal abnormality, 47,XY+21 (Trisomy-21).
Besides this two cases of Wilms tumor one is 1.5 year male showing loss of “Y” chromosome i.e. karyotype with 45, XO, and another female of 4 year showing reciprocal translocation between D/G group chromosome-15 and 21 i.e. t (15;21)(p13;p13). The present findings reveal that numerical chromosome abnormalities were more frequent than structural variations in Wilms’ tumor. Statistical analysis were carried out between total number of normal and abnormal karyotypes in the cases of Wilms’ tumor which showing significant (p<0.012) difference with variations in the values of Confidence Interval (C.I.) at 95% interval 0.0016-0.4641 and odd ratio (0.027) in trisomy-21. The variation in the frequency of karyotypic pattern with loss of chromosome showing monosomy of 21,22 and Y-chromosome, while 5% cases shows either both gain of chromosome-7 and 17, (Trisomy-9,17) or loss of chromosome i.e. monosomy of 11,18. Similar, frequency (5%) were shows structural variation (chromatid breakage) in long arm of chromosome -6 and 16 i.e. 6q21.2 and 16q23, while appearance of “ring Y” chromosome in Wilms tumors. Statistical analysis again shows the significant difference (p<0.030) with variation in the value of Confidence Interval (C.I.) at 95% interval 0.0024-0.746 and odd ratio (0.0424) in monosomy and controls. Similarly, highly significant (p<0.001) difference were observed between total structural chromosomal vs. numerical chromosomal variations with C.I. values at 95% interval 0.0365-0.2392 and odd ratio (0.934), and dicentric chromosome (46,XYdic.15) was also observed in two (2.5%) cases of Wilms tumor [29].
Discussion
The spectrums of Chromosomal Abnormalities (CAs) are the important component of tumor biology. Wilms tumor is the common embryonic solid tumor of kidney belongs to paediatric age group. Cytogenetics studies of Wilms tumors have revealed a variety of non-random distribution of trisomy of chromosome 6,7,8,12 and 18. Present cytogenetic study on Wilms tumor showing significant variation in the frequency (%) of both structural and numerical chromosomal aberrations with maximum frequency (27.5%) were observed in the karyotype having 47,XY+21 configuration which was further confirmed by FISH analysis using specific probe of DNA having 220 kb. Interestingly, these cases of Wilms tumors showing lack of clinical feature or phenotypic appearance of Down syndrome but after significant (p<0.012) statistical evaluation of involvement of extra copy of chromosome-21 confirming the “high risk” of association of Wilms tumors with Down syndrome reporting first time in eastern part of India. These observations also suggest that the existence of chromosome 21 in association with leukaemogenic genes and tumor suppression genes are involved in regulating development of Wilms tumors. The loss of heterozygocity involving two loci on 11p13 and 11p15 during development of tumors progression has been well documented, but the loss of genetic material involving two new break points (loci) mapped on 6q21.2 and 16q23 has been not been documented earlier consider as “hot spot” in Wilms tumor and cancer predisposition as reported earlier by the same author.
Although, the developing “risk” of leukemia is more common as comparison to developing solid tumor in children with down syndrome.
Earlier study reveals that incidence of lymphoreticular and solid tumors has been observed reported in down syndrome cases but showing lack of increase incidence in Wilms tumors. Interestingly, the present findings revels the occurrence of non-constitutional structural and numerical variation in karyotypes in non-syndromic cases of Wilms’ tumors needs to be further validated and may represent the power full tool as genetic marker during analysis in tumor biology. This genetic diversity showing lack of correlation between syndromic and non-syndromic cases of Wilms tumors either due to different pathogenic variations or different geographical conditions. Another, syndromic case (Edward syndrome) having triosomy-18 is associated with increased frequency of nephrogenic rests in Wilms tumor and the same were observed in the present study as documented in Table 1. The individual or in combinations with spectrum of chromosomal aberrations suggesting increase mutagenic potential due to unknown factors with increase genetic heterogeneity in syndromic and nonsyndromic cases of Wilms tumors. Earlier study showing more than 20% cases having normal karyotypes (46,XY or 46,XX) in accordance to our data also shows 23% cells having normal karyotypes perhaps due to same standard cytogenetic procedure.
Table1: The (%) frequency of total karyotypic variations in the cases of Wilms’ tumor.
However, the cytogenetics study becomes relevant to understand the primary event of interchromosomal rearrangement i.e. Robertsonian translocation or reciprocal translocation during segregation of chromosome in tumor biology. Unfortunately, our findings are interesting to make new hypothesis that involvement of more than one karyotypic variation in the same case either due to loss or gain of genetic material may develop high frequency of mosaicism for developing congenital anomalies in syndromic cases in Wilms syndrome due to antenatal exposure of environmental teratogen such as exposure of arsenic might have responsible for nondisjunction event during cell division. Only 9% cases showing rearrangement of genes (breakage and reunion) and develop complex structure i.e. the origin of dicentric of chromosome-15 or reciprocal translocation t (15;21) (p13;p13) might be explain either due to different environmental conditions which have increase the genetic susceptibility or penetrance of gene in sporadic or familial (inherited) cases of Wilms tumors. The mechanism of translocation between chromosome-15 and 21 perhaps is due to the loss of short arm of chromosome having palindromic DNA sequences leading to association between D/G groups. The another interesting findings are the appearance of rearrangement of the genes after breakage and reunion, in “ring” of Y and “loss” of Y in the karyotypes also confirm the linkage of sex- chromosome genes in familial cases of Wilms tumor, although the mode of transmission (inheritance) from parent to offspring is not clear due to lack of pedigree analysis. The data of Y chromosome in the present study on Wilms’ tumor become relevant and inaccodance with earlier findings. The molecular mechanism of Wilms’ tumour are highly complex due to genetic heterogeneity or loss of heterozygocity of these loci (11p13 and 11p15) which might have increase genetic susceptibility to kindred’s of the family of Wilms’ tumors.
However, identification of new loci assigned on different euchromatic regions of chromosome 6q21.2 and 16q23 consider as “Hot Spots” or as “tumor marker” which might have increased risk of developing congenital anomalies in solid tumors such as Wilms tumors [29].
Conclusion
The genetic diversity based on non-random distribution of chromosome aberrations between syndromic and non-syndromic cases of Wilms tumors with new findings such as origin of ring-Y and loss of Y in karyotypes explore the mechanism of tumorigenesis in Wilms tumor. Significant association of trisomy-21 with other chromosome abnormalities suggesting “risk factor” of mosaicism and their association with congenital anomalies in Wilms tumor. Although, the present study is quite interesting, however we are unable to identify many mutation that are still undetectable due to close proximity or in overlapping regions of loci containing small DNA (>5 kbp) or copy number variations. To answer these, further continuation of such study in different labs or to include larger sample size to make the study more significant in syndromic cases of Wilms tumors.
Acknowledgement
AKS is thankfully acknowledged to the Director AIIMS Patna, and Department of Science and Technology (DST) Government of India for providing financial assistance.
Conflict of Interest
There is no conflict of interest between the authors.
Authors Contributions
AKS, for providing karyotyping analysis and manuscript preparation, MT, RK, A, AK, CKS, for providing technical assistance while VK, PS for clinical diagnosis.
References
- Francke U, Holmes LB, Atkins L, et al. Aniridia-Wilms' tumor association: Evidence for specific deletion of 11p13. Cytogenet Cell Genet. 1979; 24(3):185‐192.
[Crossref] [Googlescholar] [Indexed]
- Riccardi VM, Sujansky E, Smith AC, et al. Chromosomal imbalance in the Aniridia-Wilms' tumor association: 11p interstitial deletion. Pediatrics. 1978; 61(4):604‐610.
[Crossref] [Googlescholar] [Indexed]
- Pettenati MJ, Haines JL, Higgins RR, et al. Wiedemann-Beckwith syndrome: Presentation of clinical and cytogenetic data on 22 new cases and review of the literature. Hum Genet. 1986; 74(2):143‐154.
[Crossref] [Googlescholar] [Indexed]
- Slater RM, de Kraker J, Voute PA, et al. A cytogenetic study of Wilms' tumor. Cancer Genet Cytogenet. 1985; 14(1-2):95‐109.
- Grundy P, Koufos A, Morgan K, et al. Familial predisposition to Wilms' tumour does not map to the short arm of chromosome 11. Nature. 1988; 336(6197):374‐376.
[Crossref] [Googlescholar] [Indexed]
- Huff V, Compton DA, Chao LY, et al. Lack of linkage of familial Wilms' tumour to chromosomal band 11p13. Nature. 1988; 336(6197):377‐378.
[Crossref] [Googlescholar] [Indexed]
- Kaneko Y, Egues MC, Rowley JD. Interstitial deletion of short arm of chromosome 11 limited to Wilms' tumor cells in a patient without aniridia. Cancer Res. 1981; 41(11 Pt 1):4577‐4578.
[Googlescholar] [Indexed]
- Yuan E, Li CM, Yamashiro DJ, et al. Genomic profiling maps loss of heterozygosity and defines the timing and stage dependence of epigenetic and genetic events in Wilms' tumors. Mol Cancer Res. 2005; 3(9):493‐502.
[Crossref] [Googlescholar] [Indexed]
- MdZin R, Murch A, Charles A. Cytogenetic findings in Wilms' tumour: A single institute study. Pathology. 2010; 42(7):643‐649.
[Crossref] [Googlescholar] [Indexed]
- Huff V. Wilms tumor genetics. Am J Med Genet. 1998; 79(4):260‐267. [Crossref] [Googlescholar] [Indexed]
- Peres EM, Savasan S, Cushing B, et al. Chromosome analyses of 16 cases of Wilms tumor: Different pattern in unfavorable histology. Cancer Genet Cytogenet. 2004; 148(1):66‐70.
[Crossref] [Googlescholar] [Indexed]
- Diller L, Ghahremani M, Morgan J, et al. Constitutional WT1 mutations in Wilms' tumor patients. J Clin Oncol. 1998; 16(11):3634‐3640.
[Crossref] [Googlescholar] [Indexed]
- Grundy PE, Telzerow PE, Breslow N, et al. Loss of heterozygosity for chromosomes 16q and 1p in Wilms' tumors predicts an adverse outcome. Cancer Res. 1994; 54(9):2331‐2333.
[Googlescholar] [Indexed]
- Ruteshouser EC, Robinson SM, Huff V. Wilms tumor genetics: Mutations in WT1, WTX, and CTNNB1 account for only about one-third of tumors. Genes Chromosomes Cancer. 2008; 47(6):461‐470.
[Crossref] [Googlescholar] [Indexed]
- Wilmore HP, White GF, Howell RT, et al. Germline and somatic abnormalities of chromosome 7 in Wilms' tumor. Cancer Genet Cytogenet. 1994; 77(2):93‐98.
[Crossref] [Googlescholar] [Indexed]
- Peier AM, Meloni AM, Erling MA, et al. Involvement of chromosome 7 in Wilms tumor. Cancer Genet Cytogenet. 1995; 79(1):92‐94.
[Crossref] [Googlescholar] [Indexed]
- Kusumakumary P, Ninan M, Chellam VG, et al. Wilms tumor in a child with Down syndrome. J Pediatr Hematol Oncol. 1995; 17(3):276.
[Googlescholar] [Indexed]
- Olson JM, Hamilton A, Breslow NE. Non-11p constitutional chromosome abnormalities in Wilms' tumor patients. Med Pediatr Oncol. 1995; 24:305-309.
[Crossref] [Googlescholar] [Indexed]
- Hasle H, Olsen JH, Nielsen J, et al. Occurrence of cancer in women with turner syndrome. Br J Cancer. 1996; 73(9):1156‐1159.
[Crossref] [Googlescholar] [Indexed]
- Seabright M. A rapid banding technique for human chromosomes. Lancet. 1971; 2(7731):971‐972.
[Crossref] [Googlescholar] [Indexed]
- Karayalcin G, Shanske A, Honigman R. Wilms' tumor in a 13-year old girl with trisomy 18. Am J Dis Child. 1981; 135(7):665‐666.
[Crossref] [Googlescholar] [Indexed]
- McCormick DP, Meyer WJ, Nesbit ME. Co-existance of Hodgkin’s disease and Down syndrome. Am J Dis Child 1971; 122:665-7. [Crossref] [Googlescholar] [Indexed]
- Saxena AK. Are fragile sites "hot-spots": a causative factor in tumor biology. J Exp Ther Oncol. 2012; 10(1):19‐29.
[Googlescholar] [Indexed]
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007; 7(4):233‐245.
[Crossref] [Googlescholar] [Indexed]
- Gliser CF, Schindler AM. Long term survival in a male with 18 trisomy syndrome and Wilms tumor. Paediatrics 1969; 44:111-116. [Crossref] [Googlescholar] [Indexed]
- Saha D. Arsenic ground water contamination in parts of middle plain water Bihar. Current Science. 2009; 97(6):753-55.
[Googlescholar] [Indexed]
- Dome JS, Coppes MJ. Recent advances in Wilms tumor genetics. Curr Opin Pediatr. 2002; 14(1):5‐11.
[Crossref] [Googlescholar] [Indexed]
- Clericuzio CL. Clinical phenotypes and Wilms tumor. Med Pediatr Oncol. 1993; 21 (3):182‐187.
[Crossref] [Googlescholar] [Indexed]
- Huff V, Reeve AE, Leppert M, et al. Nonlinkage of 16q markers to familial predisposition to Wilms' tumor Cancer Res. 1992;52(21):6117‐6120. [Crossref]
[Googlescholar] [Indexed]