Novel compound heterozygous mutations of the myosin heavy chain gene in patients with hypertrophic cardiomyopathy
- *Corresponding Author:
- Dr Pratibha Nallari
Department of Genetics, Osmania University, Jamia Osmania, Hyderabad, Telangana, 500 007 India.
Telephone: 91-8885486499
E-mail: prathinallari@yahoo.com
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http:// creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact support@pulsus.com
[ft_below_content] =>Keywords
Compound heterozygosity; Hypertrophic cardiomyopathy; Myosin heavy chain gene; Single-nucleotide polymorphism
Hypertrophic cardiomyopathy (HCM), a disease of the myocardium, is inherited in an autosomal dominant manner. It is characterized by thickening of the left/right ventricles and the interventricular septum in the absence of any external load, and has a prevalence of one in 500. HCM is heterogenous and the clinical course is extremely variable, ranging from asymptomatic to severe heart failure and sudden cardiac death.
Mutations in 14 sarcomeric and cytoskeletal genes have been implicated in HCM. The myosin binding heavy chain gene (MYH7) was the first gene to be implicated and accounts for approximately 35% to 50% of all HCM cases. The MYH7 gene is mapped to a region of chromosome 14q12 consisting of 41 exons, 38 of which encode the protein. The myosin heavy chain molecule has a heavy meromyosin (HMM) and a light meromyosin (LMM) region, coded by exons 3 to 28 (HMM) and 29 to 40 (LMM), respectively. Myosin hydrolyzes ATP and initiates conformational changes in the globular head and transmits to the rod domain resulting in sliding of the myofilaments of the sarcomeric apparatus. To date, >200 mutations, most of which are missense, have been reported. MYH7 mutations are generally associated with early onset of disease, extensive hypertrophy and a higher incidence of sudden cardiac death with severe penetrance (1-3).
We previously screened the head and the neck region (exons 3 to 26), which led to the identification of one known mutation (R870H) in four probands, nine single-nucleotide polymorphisms (SNPs) and two silent mutations (4). Because the rod portion of MYH7 coded by exons 29 to 40 belonging to the LMM region has not been characterized as much as the head domain with respect to SNPs/mutations, the present study screened the LMM region (exons 27 to 39 [13 exons]) to identify any pathogenic SNPs/variations in an Indian population.
Clinical evaluation
HCM was diagnosed by physical examination, electrocardiography, echocardiography and magnetic resonance imaging. Blood samples from 100 healthy blood donors with no history of heart disease were collected at the Osmania General Hospital, Hyderabad, India. One hundred HCM patients were referred by the cardiologist of CARE hospitals (Nampally, Banjara hills, Secunderabad, Hyderabad, India) for genomic DNA isolation and polymerase chain reaction (PCR)- based single-strand conformation polymorphism analysis. Clinical data and family history of the patients were also collected to facilitate risk stratification. The present study was approved by the Ethics Committee of the CARE hospital, and informed written consent was obtained from all volunteers, patients and their family members
Methods
Genomic DNA was extracted by rapid nonenzymatic method as described by Lahiri and Nurnberger (5). The isolated DNA was later amplified based on the primers described by Seidman < http://genetics. med.harvard.edu/~seidman/cg3/genes/MYBPC3_exons.html>.
PCR was performed in 0.2 mL tubes, each containing 100 ng of genomic DNA, 50 pmol each of forward and reverse primer, 0.5 to 1 unit of Taq DNA polymerase enzyme, 200 μM of dNTP, 1× PCR buffer and water to a final volume to 25 μL. Amplification was performed in a thermocycler (Mastergradient, Eppendorf, Germany). PCR conditions were: initial denaturation at 95°C for 3 min, followed by denaturation step at 95°C for 30 s. Annealing temperature varied from 54°C to 65°C (based on the exons) for 30 s and extension at 72°C for 1 min. A final extension step of 72°C was performed for 2 min at the end of the reaction. The amplified DNA samples were subjected to single-stranded conformation polymorphism analysis on 8% to 12% nondenaturing/native polyacrylamide gels. Samples showing abnormal bands or mobility shift were sequenced using a 3730xl DNA analyzer (Macrogen, Korea).
Insilico analysis
Insilico analysis was performed using Internet-based tools to predict the effect of the variation. The following tools were used for the analysis.
• Secondary protein structure prediction – PSIRED, a Protein Structure Prediction Server (http://bioinf.cs.ucl.ac.uk/psipred).
• Three-dimensional protein structure analysis – Raptor-X Pymol Visualizer (http://pymol.org/edu/index.php).
• Splice site analyses:
○ SPLICEPORT: (www.spliceport.cs.umd.edu)
○ Human splicing finder tool version 2.4.1 (http://www.umd.be/HSF)
○ Rainbow splicing: EBI Databases ASD alternative splicing (www.ebi.ac.uk/asd-srv/wb.cgi)
• Prediction of functional effects of human SNPs by PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2)
Results
Screening of exons 27 to 39 revealed three novel missense and one novel synonymous variations only in exon 34. Interstingly, patients with these variations also exhibited compound heterozygosity, suggesting exon 34 to be the ‘hotspot’ exon of the LMM region. The study also describes a unique case of a nine-year-old girl who was diagnosed with obstructive HCM and Noonan syndrome.
Case 1
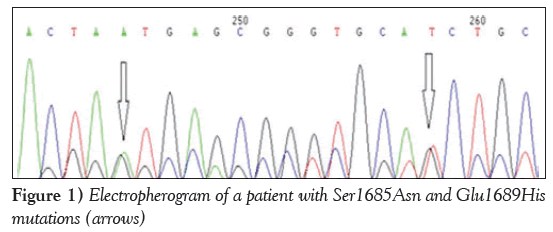
A patient (P1) exhibited two novel missense variation: a G→A transition leading to a Ser1685Asn, and a G→T transversion leading to Glu1689His (Figure 1).
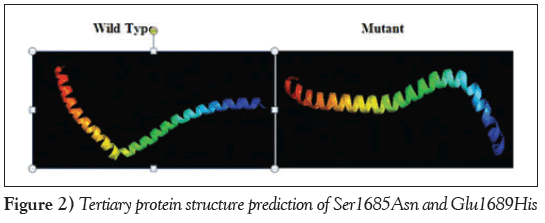
The three-dimensional protein model for the combined effect of two missense mutations was predicted using the online tool Raptor-X (http://raptorx.uchicago.edu/), which revealed a decrease in the length of the α-helix in the mutant protein, which may have an effect on protein folding and structure (Figure 2).
Human splicing finder matrices for Ser1685Asn revealed a disrupted acceptor site 10 bases upstream and a disrupted donor site three bases upstream from the start of the exon. Exonic splicing enhancer finder matrices predicted a disrupted binding site for spliceosome binding protein SRp40. The exonic splicing enhancer binding site was replaced by a silencer binding site, which could alter pre-messenger RNA splicing. For Glu1689His, the acceptor site was observed to be disrupted at nine bases upstream from the site of the variation. At the site of the variation, the splicing regulatory sequence was observed to be disrupted and replaced by a cryptic silencer binding site.
Case 2
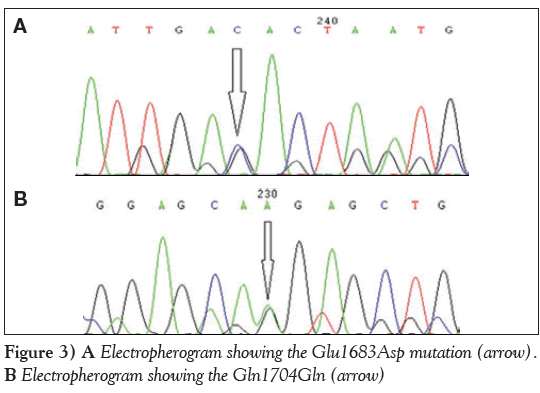
The second case of genetic compound heterozygosity with a novel G→A transition resulting in a synonymous Gln1704Gln change and G→C transversion leading to a missense Glu1683Asp change was observed in two patient samples (P2 and P3) (Figure 3A and Figure 3B). The secondary and tertiary protein structure prediction did not reveal any change in the mutant protein.
Clinical correlation
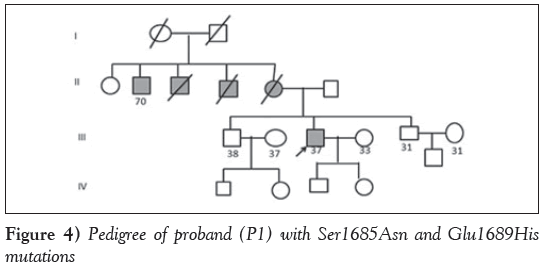
Case 1: The proband with missense mutations Ser1685Asn and Glu1689His (P1) was a 37-year-old man diagnosed with HCM with symptoms of dyspnea and occasional palpitations at 33 years of age with a strong family history of sudden cardiac death, wherein the mother and two maternal uncles died of HCM, and one uncle survived with the disease. Echocardiography revealed an intact ventricular septum (IVS) of 2.3 cm, left ventricular posterior wall (LVPW) of 1.2 cm, ejection fraction (EF) of 61% and left ventricular outflow tract (LVOT) of 45 mmHg. The genetic compound observed in the patient could have accounted for the severe penetrance (Figure 4).
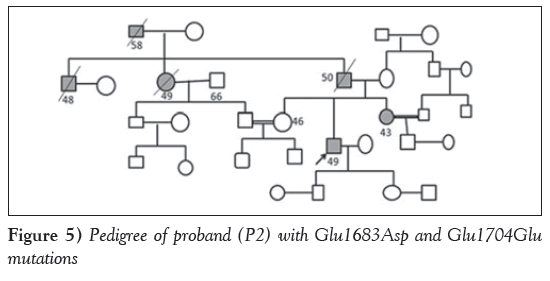
Case 2: Two patients revealed a novel missense variation Glu1683Asp and a novel synonymous variation; Glu1704Glu leading to compound heterozygosity, wherein one of the probands (P2), a 49-year-old man, was diagnosed with nonobstructive HCM at 29 years of age with presenting symptoms of palpitations, chest pain, and presyncope with hypercholesterolemia. Echocardiography revealed an IVS of 2.5 cm, an LVPW of 1.2 cm, an EF of 72% and LVOT gradient of 63 mmHg. Family history of sudden cardiac death was noted (the father, paternal uncle and aunt had died suddenly). Parental consanguinity (first cousin) was also reported in the family (Figure 5).
The second patient (P3) was a 43-year-old man diagnosed with nonobstructive HCM with symptoms of dyspnea and diabetes. Echocardiography revealed an IVS of 1.5 cm, an LVPW of 1.3 cm and EF of 78%. The proband’s father experienced sudden cardiac death while exercising.
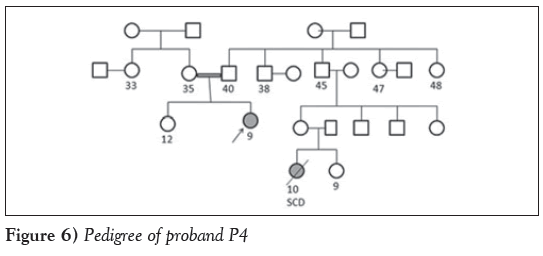
Case 3: Another unique case of a nine-year-old girl (P4) who was diagnosed with obstructive HCM and Noonan syndrome after presenting with symptoms of dyspnea, palpitation and chest pain was studied. Echocardiography revealed an IVS of 0.96 cm, an LVPW of 1.5 cm, an EF of 60% and LVOT of 92 mmHg. The parents were in a consanguineous marriage and the proband’s cousin, a 10-year-old girl, died from sudden cardiac death. Molecular analyses revealed a synonymous mutation (Gln1704Gln) in exon 34 of the MYH7 gene. Insilico analysis revealed a disrupted binding site for spliceosome protein SF2/ASF. A cryptic enhancer binding site for splicing regulatory sequence and 9G8 and Tra2-β proteins were also predicted. The silencer motif was disrupted and replaced by a cryptic enhancer binding site (Figure 6).
One of the common findings associated with these variations was the history of sudden death in the families, strong disease penetrant gene with early age at onset and compound heterozygosity, all being phenotypic hallmarks of MYH7 gene variations.
Discussion
Myosin is a highly asymmetric protein with two globular heads (S1) and a long rod domain (S2). The S1 head is the enzymatic end of the molecule and possesses the actin and ATP binding domains and is responsible for force transduction. Most of the MYH7 mutations reported are in the S1 head domain, which is the highly conserved region (6,7).
Previous reports have speculated that >30% of all HCM cases are due to mutations in MYH7, wherein most of the mutations have been observed in the head and neck regions (8). However, there have been only eight mutations reported in the rod portion of MYH7 to date, which still amount to 20% of HCM cases (9,10). We previously screened the head and the neck region (exons 3 to 26), which led to the identification of one known mutation (R870H) in four probands, nine SNPs and two silent mutations (4).
Therefore, the present study considered screening of the rod region, which revealed variations only in exon 34. Three novel missense mutations (Ser1685Asn, Glu1689His and Glu1683Asp) and one novel synonymous (Gln1704Gln) were observed, along with three cases of compound heterozygosity, revealing the conserved nature of the LMM region (rod) suggesting it to be the ‘hotspot’. This was in accordance with a previous study by Tanjore et al (4) who also reported only 4.2% of MYH7 mutations.
A very low rate of MYH7 mutations has also been observed in a Finnish population (5.6% of HCM cases) (11). A study by Van Driest et al (12) found that 15% of patients harboured MYH7 mutations in a large Caucasian cohort. In contrast, a comprehensive analysis of 197 unrelated patients of a French cohort revealed 25% of MYH7 mutations (10). This difference could suggest a strong selective pressure against nucleotide substitutions that may predispose to the development of HCM. This finding further strengthens the genetic diversity of various populations, and particularly in the Indian population.
Given that the rod portion of MYH7 has been less well characterized than the head domain, it is possible that mutations in the rod region represent nonpathogenic SNPs, disease modifiers or SNPs linked to mutations in a neighbouring gene. Any variations in these sarcomeric genes can result in disarray and hypertrophy. High-throughput sequence analysis involving larger cohorts of healthy individuals will provide additional data as to the prevalence and spectrum of nonpathogenic MYH7 variants.
In the present study, screening of the LMM region revealed novel variations in only exon 34, indicating that it could be a hotspot region in the rod domain and most of the patients exhibited compound heterozygosity, which may account for the increased penetrance and disease phenotype. Interestingly, the patients with putative rod-domain mutations presented with a clinical phenotype indistinguishable from those with mutations involving the head and neck regions of MYH7. Therefore, screening of the entire MYH7 gene, involving the head, neck and rod regions, is warranted in genetic diagnosis of cardiomyopathy.
Conclusion
The present study considered screening of the LMM region constituting the rod portion of the MYH7 gene, which revealed three novel missense and one novel synonymous variation in exon 34, indicating it to be a hotspot exon of the LMM region of MYH7 gene. The present study emphasizes the importance of LMM region of MYH7 gene, suggesting screening of the entire gene for HCM diagnosis.
Acknowledgements
The contribution of all the authors is acknowledged. The authors thank Indian Council of Medical Research and Department of Science and Technology, New Delhi for the financial assistance.
Disclosures: The authors have no financial disclosures or conflicts of interest to declare.
References
- Niimura H, Bachinski LL, Sangwatanaroj S, et al. Mutations in the gene for cardiac myosinbinding protein c and late-onset familial hypertrophic cardiomyopathy. N Engl J Med 1998;338:1248-57.
- Charron P, Dubourg O, Desnos M, et al. Genotype-phenotype correlations in familial hypertrophic cardiomyopathy. A comparison between mutations in the cardiac protein-C and the beta-myosin heavy chain genes. Eur Heart J 1998;19:139-45.
- Redwood CS, Moolman-Smook JC, Watkins H. Properties of mutant contractile proteins that cause hypertrophic cardiomyopathy. Cardiovasc Res 1999;44:20-36.
- Tanjore RR, Sikindlapuram AD, Calambur N, Thakkar B, Kerkar PG, Nallari P. Genotype-phenotype correlation of R870H mutation in hypertrophic cardiomyopathy. Clin Genet 2006;69:434-6.
- Lahiri D, Nurnberger J. A rapid non enzymatic method for preparation of HMW DNA from blood for RFLP studies. Nucleic Acid Res1991:19:5444.
- Warrick HM, Spudich JA. Mysoin structure and function in cell motility. Ann Rev Cell Biol 1987;3:379-421.
- Mornet D, Bonet A, Audemard E, Bonicel J. Functional sequences of Myosin head. J Musc Cell Mot 1989;10:10-24.
- Seidman JG, Seidman C. The genetic basis for cardiomyopathy: From mutation identification to mechanistic paradigms. Cell 2001;104:557-67.
- Blair E, Redwood C, Watkins H, et al. Ascertainment strategies and genotype: Phenotype correlations in hypertrophic cardiomyopathy: Response. Circulation 2003;108:24e-5e.
- Richard P, Charron P, Carrier L, et al. Hypertrophic cardiomyopathy: Distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy.Circulation 2003;107:2227-32.
- Jeschke B, Uhl K, Weist B, et al. A high risk phenotype of hypertrophic cardiomyopathy associated with a compound genotype of two mutated beta-myosin heavy chain genes. Hum Genet 1998;102:299-304.
- Van Driest SL, Varile VC, Ommen SR, et al. Myosin binding protein C mutation and compound heterozygosites in hypertrophic cardiomyopathy. J AM Coll Cardiol 2004;44:1903-10.
- *Corresponding Author:
- Dr Pratibha Nallari
Department of Genetics, Osmania University, Jamia Osmania, Hyderabad, Telangana, 500 007 India.
Telephone: 91-8885486499
E-mail: prathinallari@yahoo.com
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http:// creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact support@pulsus.com
Abstract
background: Hypertrophic cardiomyopathy (HCM) is a multifactorial disorder, with mutations implicated in 14 sarcomeric and cytoskeletal genes, leading to genotypic and phenotypic heterogeneity, and a challenging genetic and clinical diagnosis. The genetic characteristics of HCM have been studied for more than two decades in various ethnic and racial groups, and many novel genetic variations have been reported. The myosin heavy chain gene is the most heavily implicated gene in HCM, with >200 reported mutations, the majority of which have been found in the head-rod junction. The rod portion of MYH7, coded by exons 29 to 40 and belonging to the light meromyosin (LMM) region, has not been characterized to the same extent as the head domain with respect to single-nucleotide polymorphisms (SNPs)/mutations.
OBJECTIVE: To screen the conserved LMM region, constituting exons 27 to 39 (13 exons), to identify any pathogenic SNPs/variations in this region in a population of Indian patients.
Methods: Molecular screening was performed by polymerase chain reaction-based single-strand conformation polymorphism analysis in 100 control individuals and 100 HCM patients. The variations were confirmed by sequencing. Insilico analysis was performed to analyze the effect of the respective variations.
Results : Screening of exons 27 to 39 revealed three novel missense variations and one novel synonymous variation in exon 34. Interestingly, patients with these variations also exhibited compound heterozygosity, indicating exon 34 to be the ‘hotspot’ exon of the LMM region.
Conclusion: The results of the present study emphasize the importance of the LMM (rod) region of the MYH7 gene and suggest that variations in the conserved region are likely to be more pathogenic, making screening of the entire gene for HCM diagnosis mandatory.
-Keywords
Compound heterozygosity; Hypertrophic cardiomyopathy; Myosin heavy chain gene; Single-nucleotide polymorphism
Hypertrophic cardiomyopathy (HCM), a disease of the myocardium, is inherited in an autosomal dominant manner. It is characterized by thickening of the left/right ventricles and the interventricular septum in the absence of any external load, and has a prevalence of one in 500. HCM is heterogenous and the clinical course is extremely variable, ranging from asymptomatic to severe heart failure and sudden cardiac death.
Mutations in 14 sarcomeric and cytoskeletal genes have been implicated in HCM. The myosin binding heavy chain gene (MYH7) was the first gene to be implicated and accounts for approximately 35% to 50% of all HCM cases. The MYH7 gene is mapped to a region of chromosome 14q12 consisting of 41 exons, 38 of which encode the protein. The myosin heavy chain molecule has a heavy meromyosin (HMM) and a light meromyosin (LMM) region, coded by exons 3 to 28 (HMM) and 29 to 40 (LMM), respectively. Myosin hydrolyzes ATP and initiates conformational changes in the globular head and transmits to the rod domain resulting in sliding of the myofilaments of the sarcomeric apparatus. To date, >200 mutations, most of which are missense, have been reported. MYH7 mutations are generally associated with early onset of disease, extensive hypertrophy and a higher incidence of sudden cardiac death with severe penetrance (1-3).
We previously screened the head and the neck region (exons 3 to 26), which led to the identification of one known mutation (R870H) in four probands, nine single-nucleotide polymorphisms (SNPs) and two silent mutations (4). Because the rod portion of MYH7 coded by exons 29 to 40 belonging to the LMM region has not been characterized as much as the head domain with respect to SNPs/mutations, the present study screened the LMM region (exons 27 to 39 [13 exons]) to identify any pathogenic SNPs/variations in an Indian population.
Clinical evaluation
HCM was diagnosed by physical examination, electrocardiography, echocardiography and magnetic resonance imaging. Blood samples from 100 healthy blood donors with no history of heart disease were collected at the Osmania General Hospital, Hyderabad, India. One hundred HCM patients were referred by the cardiologist of CARE hospitals (Nampally, Banjara hills, Secunderabad, Hyderabad, India) for genomic DNA isolation and polymerase chain reaction (PCR)- based single-strand conformation polymorphism analysis. Clinical data and family history of the patients were also collected to facilitate risk stratification. The present study was approved by the Ethics Committee of the CARE hospital, and informed written consent was obtained from all volunteers, patients and their family members
Methods
Genomic DNA was extracted by rapid nonenzymatic method as described by Lahiri and Nurnberger (5). The isolated DNA was later amplified based on the primers described by Seidman < http://genetics. med.harvard.edu/~seidman/cg3/genes/MYBPC3_exons.html>.
PCR was performed in 0.2 mL tubes, each containing 100 ng of genomic DNA, 50 pmol each of forward and reverse primer, 0.5 to 1 unit of Taq DNA polymerase enzyme, 200 μM of dNTP, 1× PCR buffer and water to a final volume to 25 μL. Amplification was performed in a thermocycler (Mastergradient, Eppendorf, Germany). PCR conditions were: initial denaturation at 95°C for 3 min, followed by denaturation step at 95°C for 30 s. Annealing temperature varied from 54°C to 65°C (based on the exons) for 30 s and extension at 72°C for 1 min. A final extension step of 72°C was performed for 2 min at the end of the reaction. The amplified DNA samples were subjected to single-stranded conformation polymorphism analysis on 8% to 12% nondenaturing/native polyacrylamide gels. Samples showing abnormal bands or mobility shift were sequenced using a 3730xl DNA analyzer (Macrogen, Korea).
Insilico analysis
Insilico analysis was performed using Internet-based tools to predict the effect of the variation. The following tools were used for the analysis.
• Secondary protein structure prediction – PSIRED, a Protein Structure Prediction Server (http://bioinf.cs.ucl.ac.uk/psipred).
• Three-dimensional protein structure analysis – Raptor-X Pymol Visualizer (http://pymol.org/edu/index.php).
• Splice site analyses:
○ SPLICEPORT: (www.spliceport.cs.umd.edu)
○ Human splicing finder tool version 2.4.1 (http://www.umd.be/HSF)
○ Rainbow splicing: EBI Databases ASD alternative splicing (www.ebi.ac.uk/asd-srv/wb.cgi)
• Prediction of functional effects of human SNPs by PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2)
Results
Screening of exons 27 to 39 revealed three novel missense and one novel synonymous variations only in exon 34. Interstingly, patients with these variations also exhibited compound heterozygosity, suggesting exon 34 to be the ‘hotspot’ exon of the LMM region. The study also describes a unique case of a nine-year-old girl who was diagnosed with obstructive HCM and Noonan syndrome.
Case 1
A patient (P1) exhibited two novel missense variation: a G→A transition leading to a Ser1685Asn, and a G→T transversion leading to Glu1689His (Figure 1).
The three-dimensional protein model for the combined effect of two missense mutations was predicted using the online tool Raptor-X (http://raptorx.uchicago.edu/), which revealed a decrease in the length of the α-helix in the mutant protein, which may have an effect on protein folding and structure (Figure 2).
Human splicing finder matrices for Ser1685Asn revealed a disrupted acceptor site 10 bases upstream and a disrupted donor site three bases upstream from the start of the exon. Exonic splicing enhancer finder matrices predicted a disrupted binding site for spliceosome binding protein SRp40. The exonic splicing enhancer binding site was replaced by a silencer binding site, which could alter pre-messenger RNA splicing. For Glu1689His, the acceptor site was observed to be disrupted at nine bases upstream from the site of the variation. At the site of the variation, the splicing regulatory sequence was observed to be disrupted and replaced by a cryptic silencer binding site.
Case 2
The second case of genetic compound heterozygosity with a novel G→A transition resulting in a synonymous Gln1704Gln change and G→C transversion leading to a missense Glu1683Asp change was observed in two patient samples (P2 and P3) (Figure 3A and Figure 3B). The secondary and tertiary protein structure prediction did not reveal any change in the mutant protein.
Figure 3: A Electropherogram showing the Glu1683Asp mutation (arrow). B Electropherogram showing the Gln1704Gln (arrow)
Clinical correlation
Case 1: The proband with missense mutations Ser1685Asn and Glu1689His (P1) was a 37-year-old man diagnosed with HCM with symptoms of dyspnea and occasional palpitations at 33 years of age with a strong family history of sudden cardiac death, wherein the mother and two maternal uncles died of HCM, and one uncle survived with the disease. Echocardiography revealed an intact ventricular septum (IVS) of 2.3 cm, left ventricular posterior wall (LVPW) of 1.2 cm, ejection fraction (EF) of 61% and left ventricular outflow tract (LVOT) of 45 mmHg. The genetic compound observed in the patient could have accounted for the severe penetrance (Figure 4).
Case 2: Two patients revealed a novel missense variation Glu1683Asp and a novel synonymous variation; Glu1704Glu leading to compound heterozygosity, wherein one of the probands (P2), a 49-year-old man, was diagnosed with nonobstructive HCM at 29 years of age with presenting symptoms of palpitations, chest pain, and presyncope with hypercholesterolemia. Echocardiography revealed an IVS of 2.5 cm, an LVPW of 1.2 cm, an EF of 72% and LVOT gradient of 63 mmHg. Family history of sudden cardiac death was noted (the father, paternal uncle and aunt had died suddenly). Parental consanguinity (first cousin) was also reported in the family (Figure 5).
The second patient (P3) was a 43-year-old man diagnosed with nonobstructive HCM with symptoms of dyspnea and diabetes. Echocardiography revealed an IVS of 1.5 cm, an LVPW of 1.3 cm and EF of 78%. The proband’s father experienced sudden cardiac death while exercising.
Case 3: Another unique case of a nine-year-old girl (P4) who was diagnosed with obstructive HCM and Noonan syndrome after presenting with symptoms of dyspnea, palpitation and chest pain was studied. Echocardiography revealed an IVS of 0.96 cm, an LVPW of 1.5 cm, an EF of 60% and LVOT of 92 mmHg. The parents were in a consanguineous marriage and the proband’s cousin, a 10-year-old girl, died from sudden cardiac death. Molecular analyses revealed a synonymous mutation (Gln1704Gln) in exon 34 of the MYH7 gene. Insilico analysis revealed a disrupted binding site for spliceosome protein SF2/ASF. A cryptic enhancer binding site for splicing regulatory sequence and 9G8 and Tra2-β proteins were also predicted. The silencer motif was disrupted and replaced by a cryptic enhancer binding site (Figure 6).
One of the common findings associated with these variations was the history of sudden death in the families, strong disease penetrant gene with early age at onset and compound heterozygosity, all being phenotypic hallmarks of MYH7 gene variations.
Discussion
Myosin is a highly asymmetric protein with two globular heads (S1) and a long rod domain (S2). The S1 head is the enzymatic end of the molecule and possesses the actin and ATP binding domains and is responsible for force transduction. Most of the MYH7 mutations reported are in the S1 head domain, which is the highly conserved region (6,7).
Previous reports have speculated that >30% of all HCM cases are due to mutations in MYH7, wherein most of the mutations have been observed in the head and neck regions (8). However, there have been only eight mutations reported in the rod portion of MYH7 to date, which still amount to 20% of HCM cases (9,10). We previously screened the head and the neck region (exons 3 to 26), which led to the identification of one known mutation (R870H) in four probands, nine SNPs and two silent mutations (4).
Therefore, the present study considered screening of the rod region, which revealed variations only in exon 34. Three novel missense mutations (Ser1685Asn, Glu1689His and Glu1683Asp) and one novel synonymous (Gln1704Gln) were observed, along with three cases of compound heterozygosity, revealing the conserved nature of the LMM region (rod) suggesting it to be the ‘hotspot’. This was in accordance with a previous study by Tanjore et al (4) who also reported only 4.2% of MYH7 mutations.
A very low rate of MYH7 mutations has also been observed in a Finnish population (5.6% of HCM cases) (11). A study by Van Driest et al (12) found that 15% of patients harboured MYH7 mutations in a large Caucasian cohort. In contrast, a comprehensive analysis of 197 unrelated patients of a French cohort revealed 25% of MYH7 mutations (10). This difference could suggest a strong selective pressure against nucleotide substitutions that may predispose to the development of HCM. This finding further strengthens the genetic diversity of various populations, and particularly in the Indian population.
Given that the rod portion of MYH7 has been less well characterized than the head domain, it is possible that mutations in the rod region represent nonpathogenic SNPs, disease modifiers or SNPs linked to mutations in a neighbouring gene. Any variations in these sarcomeric genes can result in disarray and hypertrophy. High-throughput sequence analysis involving larger cohorts of healthy individuals will provide additional data as to the prevalence and spectrum of nonpathogenic MYH7 variants.
In the present study, screening of the LMM region revealed novel variations in only exon 34, indicating that it could be a hotspot region in the rod domain and most of the patients exhibited compound heterozygosity, which may account for the increased penetrance and disease phenotype. Interestingly, the patients with putative rod-domain mutations presented with a clinical phenotype indistinguishable from those with mutations involving the head and neck regions of MYH7. Therefore, screening of the entire MYH7 gene, involving the head, neck and rod regions, is warranted in genetic diagnosis of cardiomyopathy.
Conclusion
The present study considered screening of the LMM region constituting the rod portion of the MYH7 gene, which revealed three novel missense and one novel synonymous variation in exon 34, indicating it to be a hotspot exon of the LMM region of MYH7 gene. The present study emphasizes the importance of LMM region of MYH7 gene, suggesting screening of the entire gene for HCM diagnosis.
Acknowledgements
The contribution of all the authors is acknowledged. The authors thank Indian Council of Medical Research and Department of Science and Technology, New Delhi for the financial assistance.
Disclosures: The authors have no financial disclosures or conflicts of interest to declare.
References
- Niimura H, Bachinski LL, Sangwatanaroj S, et al. Mutations in the gene for cardiac myosinbinding protein c and late-onset familial hypertrophic cardiomyopathy. N Engl J Med 1998;338:1248-57.
- Charron P, Dubourg O, Desnos M, et al. Genotype-phenotype correlations in familial hypertrophic cardiomyopathy. A comparison between mutations in the cardiac protein-C and the beta-myosin heavy chain genes. Eur Heart J 1998;19:139-45.
- Redwood CS, Moolman-Smook JC, Watkins H. Properties of mutant contractile proteins that cause hypertrophic cardiomyopathy. Cardiovasc Res 1999;44:20-36.
- Tanjore RR, Sikindlapuram AD, Calambur N, Thakkar B, Kerkar PG, Nallari P. Genotype-phenotype correlation of R870H mutation in hypertrophic cardiomyopathy. Clin Genet 2006;69:434-6.
- Lahiri D, Nurnberger J. A rapid non enzymatic method for preparation of HMW DNA from blood for RFLP studies. Nucleic Acid Res1991:19:5444.
- Warrick HM, Spudich JA. Mysoin structure and function in cell motility. Ann Rev Cell Biol 1987;3:379-421.
- Mornet D, Bonet A, Audemard E, Bonicel J. Functional sequences of Myosin head. J Musc Cell Mot 1989;10:10-24.
- Seidman JG, Seidman C. The genetic basis for cardiomyopathy: From mutation identification to mechanistic paradigms. Cell 2001;104:557-67.
- Blair E, Redwood C, Watkins H, et al. Ascertainment strategies and genotype: Phenotype correlations in hypertrophic cardiomyopathy: Response. Circulation 2003;108:24e-5e.
- Richard P, Charron P, Carrier L, et al. Hypertrophic cardiomyopathy: Distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy.Circulation 2003;107:2227-32.
- Jeschke B, Uhl K, Weist B, et al. A high risk phenotype of hypertrophic cardiomyopathy associated with a compound genotype of two mutated beta-myosin heavy chain genes. Hum Genet 1998;102:299-304.
- Van Driest SL, Varile VC, Ommen SR, et al. Myosin binding protein C mutation and compound heterozygosites in hypertrophic cardiomyopathy. J AM Coll Cardiol 2004;44:1903-10.