Pharmacogenomics and/or therapeutic drug monitoring in cancer chemotherapy are used independently or together for dose optimization
Received: 04-Aug-2018 Accepted Date: Sep 03, 2018; Published: 12-Sep-2018
Citation: Desoky E. Pharmacogenomics and/or therapeutic drug monitoring in cancer chemotherapy are used independently or together for dose optimization. Clin Pharmacol Toxicol Res. 2018;1(2):19-22.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Due to the wide interindividual variability in pharmacokinetics and therapeutic response of cancer patients towards anticancer drugs, pharmacogenomic-assisted dose optimization and/or application of therapeutic drug monitoring service could be of benefit. For some drugs like irinotecan used in solid tumors, dose optimization may be achieved if the oncologist depends on pharmacogenomic approach. On the other hand, dose adjustment of some other anticancer drugs like tamoxifen used in breast cancer can be affected more by application of therapeutic drug monitoring of its active metabolite; endoxifen.
Keywords
Pharmacogenomics, Therapeutic drug monitoring, Irinotecan, Tamoxifen
Introduction
Despite the notable advances in treatments used in oncology, interindividual variabilities in drug response, and/or side effects may limit the use of chemotherapy protocols [1]. The drugs used in oncology require often a dosage adjustment to limit the toxic effects while maintaining optimal efficiency [2].
Irinotecan in solid tumors like colon and lung cancers and tamoxifen in estrogen receptor (ER)-positive breast cancer are widely used. The clinical response, efficacy and toxicity of both drugs are variable and may be related to pharmacogenomic (PG) aspect and/or biotransformation of the drugs and their active metabolites.
Implementation of the tumor pharmacogenomic in routine clinical practice, might be of interest as it could predict the future of oncology [3]. Many factors complicate the issue of dose optimization in cancer chemotherapy including availability of different therapeutic protocols, wide interindividual variability in PG and pharmacokinetics (PK) of anticancer drugs and tumor resistance to drug therapy [4,5].
In this minireview, we have selected irinotecan and tamoxifen as two examples of anticancer drugs to discuss whether PG aspect can play a definite role in dose optimization, clinical outcome and drug toxicity or application of therapeutic drug monitoring could be more reliable especially for tamoxifen drug and its active metabolite, endoxifen.
I-Irinotecan
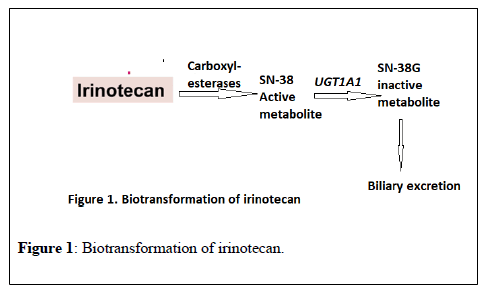
Irinotecan is an antineoplastic agent widely used in colon and lung cancers. It is bio transformed in the liver by carboxylesterase 2 to 7- ethyl-10-hydroxycamptothecin (SN-38), an active metabolite 100-1000 times more cytotoxic than the parent compound. SN38 is metabolized via glucuronidation by a UDP-glucuronosyltransferase (UGT) enzyme encoded by the UGT1A1 gene to an inactive compound, SN38 G, more easily eliminated in bile and urine (Figure 1) [6]. The risk of irinotecan toxicity increases with genetic variants associated with reduced UGT enzyme activity. Therefore, UGT1A1 may be a predictor for irinotecan-induced toxicity. More than sixty variants of UGT1A1 have been reported and UGT1A1*1 is the wild-type allele associated with normal enzyme activity. On the other hand, the most common variant (mutant) alleles related to irinotecan-induced toxicity are UGT1A1*28 and UGT1A1*6 [7].
The frequency of UGT1A1*28, is about 45% in African Americans, up to 40% in Caucasians but less common in Asian population (about 10%) [8-10].
On the other hand, the variant allele, UGT1A1*6, is more prevalent in Asian populations, with an allele frequency up to 40% [11,12], however, is rare in Caucasians [13].
The reduced UGT enzyme activity with the genetic variants UGT1A1*28 increases the risk of neutropenia from treatment with irinotecan due to reduced excretion of irinotecan metabolites, which leads to increased active irinotecan metabolites in the blood. Approximately 10% of North Americans carry 2 copies of the UGT1A1*28 allele (homozygous, UGT1A1 *28/*28), and are more likely to develop neutropenia following irinotecan therapy [14,15]. The FDA-approved drug label for irinotecan states that “when administered as a single-agent, a reduction in the starting dose by at least one level of irinotecan hydrochloride injection should be considered for patients known to be homozygous for the UGT1A1*28 allele [16]. Furthermore, Innocenti et al in 2013 demonstrated the interest of genotyping of UGT1A1*28 in the dose setting of irinotecan. Patients with advanced solid tumors exposed to irinotecan were stratified by the *1/*1, *1/*28, and *28/*28 genotypes. In the group with the *1/*1 genotype, the maximal tolerated dose (MTD) of the drug was 850 mg, the group with the *1/*28 genotype, the MTD was 700 mg while the group of patients with the *28/*28 genotype, the MTD was 400 mg. While the irinotecan area under plasma concentration curve (AUC) was correlated with the actual dose given in each group in a linear way, the different doses of irinotecan in the different three groups with different variants of UGT1A1*28 resulted in similar SN-38 areas under the curve [17]. The study concluded that UGT1A1*28 genotype can be used to individualize dosing of irinotecan and additional studies should evaluate the effect of genotype-guided dosing on efficacy in patients receiving irinotecan.
As regards irinotecan-induced diarrhea as another toxicity, UGT1A1*28/*28 genotype had a higher risk of diarrhea than those with UGT1A1*1/*1. The data of 25 studies were used to analyze the association between diarrhea and UGT1A1*28 polymorphism whereas those with UGT1A1*1/*28 had intermediate risk [7,18].
Analysis of the association between UGT1A1*6 polymorphism and neutropenia showed that the risk of that toxicity was higher in AA genotype than GG genotype whereas heterozygotes had intermediate risk [19,20].
The risk of irinotecan-induced diarrhea in patients carrying UGT1A1*6 gene variants has been reported higher in patients carrying AA genotype than GG genotype whereas heterozygotes showed intermediate risk [21,22].
According to the above, UGT1A1 genotyping can be considered as a straightforward approach for irinotecan dose individualization to achieve therapeutic targets and reduce adverse effects while application of TDM of the drug is not at the same beneficial level.
II-Tamoxifen
In the Western world, breast cancer is the most commonly diagnosed malignancy in women representing the second leading cause of cancer deaths. This type of cancer is often dependent on estrogen in growth and anti-estrogen medications are commonly used in treatment [23].
Tamoxifen is a selective estrogen receptor modulator (SERM) that has anti-estrogenic effect on the mammary gland and is used in hormone receptor-positive breast cancer in premenopausal women as a long-term adjuvant therapy showing a 50% reduction in annual recurrence rate and a 30% reduction in breast cancer mortality following 5 years of treatment [24,25].
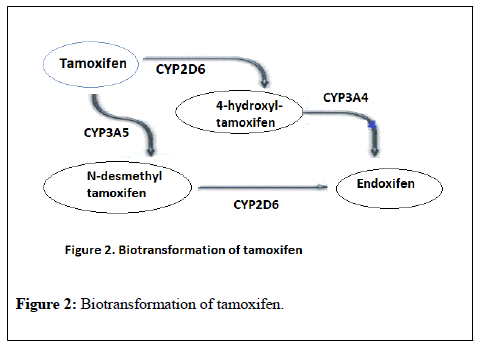
Tamoxifen is extensively metabolized in the liver mainly by CYP2D6 and CYP3A4 into a range of active and inactive metabolites (Figure 2).
4-hydroxytamoxifen (4OH-tamoxifen) and N-desmethyl-4-hydroxytamoxifen (endoxifen), both have an affinity for the estrogen receptor that markedly exceeds that of tamoxifen itself. Endoxifen is considered the main active metabolite, though it has substantially lower steady-state concentrations than tamoxifen itself in blood, but its affinity for the estrogen receptor is at least 100-fold higher than the parent drug [26].
CYP2D6 is highly polymorphic and is considered as the rate limiting enzyme in tamoxifen to endoxifen bioactivation. The metabolic activity of that enzyme can be predicted from CYP2D6 genotype, enabling classification of patients into poor (PM), intermediate (IM), extensive (EM), or ultra-rapid (UM) metabolizer phenotypes [27,28]. The high variability in endoxifen concentrations among patients ranging from 5 nM to 100 nM is thought to be derived from polymorphism in CY2D6 [29].
The importance of CYP2D6 in tamoxifen metabolism and subsequent endoxifen formation has provided logical rationale for the hypothesis that CYP2D6 genotype correlates with tamoxifen efficacy, disease outcomes, and prediction of the risk of recurrence [30,31].
However, other studies failed to provide conclusive evidence for recommending CYP2D6 genotyping as a predictive marker of tamoxifen efficacy [32,33].
Endoxifen threshold which has a therapeutic effect was reported as 5.97 ng/ml (=16.0 nM) and Patients with low endoxifen blood concentrations less than this threshold value may experience less tamoxifen efficacy and may require a tamoxifen dose higher than the standard 20 mg once daily dose [34]. The low endoxifen level achieved with administration of fixed dose of tamoxifen (20 mg/day) may be multifactorial, and CYP2D6 genotype is one of them. Other factors may include, concomitant CYP2D6 inhibitor use, poor adherence (non-compliance) and impaired absorption. One or more of these factors were identified in 48% of those with endoxifen <5.9 ng/ml. Therefore, it has been reported that 50% of low endoxifen subjects could not be explained by CYP2D6 genotype or use of inhibitory medications but even more these subjects may not have obvious causes identified [35].
Although the PM CYP2D6 genotype give insight to low endoxifen levels, some PM individuals may have baseline endoxifen >5.9 ng/ml and others could achieve that level with tamoxifen dose escalation. Conversely, UM/EM and IM patients may have endoxifen levels <5.9 ng/ml even when the use of concomitant inhibitors of CYP2D6 is excluded. Therefore, determining the CYP2D6 genotype is likely not enough to predict the concentration of the active metabolite endoxifen [36].
On the other hand, endoxifen concentration monitoring has been suggested to play a key role in modulating tamoxifen's clinical effectiveness. Based pharmacokinetic-pharmacodynamic modeling, optimal tumor growth inhibition (TGI) was predicted for patients attaining endoxifen concentrations >40 nM, while <15 nM endoxifen was predicted to achieve sub-optimal (83%) TGI [37]. In a large retrospective study on endoxifen threshold in estrogen receptor-positive breast cancer patients administered tamoxifen, patients with an endoxifen serum concentration higher than 5.9 ng/mL had a 26% lower recurrence rate than patients with a lower endoxifen serum concentration [38]. Another study reported that patients with <14 nM endoxifen had significantly short distant relapse-free survival as compared to patients with levels >35 nM [31]. Dose escalation of tamoxifen based on baseline endoxifen concentration rather than CYP2D6 genotype has made sound in PM patients with subtherapeutic level of endoxifen (less than 5.6 ng/ml). Increase the dose from 20 mg to 30-40 mg daily elevated endoxifen level significantly to be similar to normal metabolizers averages, without any noted increase in adverse events in CYP2D6 PM subjects [34,39,40].
Conclusion
In cancer chemotherapy, therapeutic efficacy and reduction of toxicity are two main therapeutic goals for oncologists. Pharmacogenomics and/or therapeutic drug monitoring are two pharmacological tools that can help dose optimization to achieve these goals.
Irinotecan used in colon and lung cancers is an example of anticancer agents where pharmacogenomics could be utilized during its use. UGT1A1*28 and UGT1A1*6 polymorphisms may help in predicting severe drug toxicity, such as neutropenia and diarrhea, and determining the maximal tolerated dose of the drug.
Tamoxifen is used as a long-term adjuvant therapy in premenopausal women with cancer breast. The biotransformation of the drug to its active metabolite; endoxifen, by the highly polymorphic CYP2D6 enzyme drew the attention for the utility of testing the genotyping profile of that enzyme in patients receiving the drug. However, it is unclear whether there is an association between CYP2D6 genotype and tamoxifen dosage and efficacy. Therefore, additional research is needed to correlate genetic data with individualization of drug dosage. On the other hand, therapeutic drug monitoring of endoxifen concentration in the blood seems promising to be relied upon as a surrogate marker of adequate tamoxifen dosing. Further studies on a large scale are suggested to identify an optimal endoxifen concentration target and to validate its correlation with treatment efficacy.
REFERENCES
- Gervasini G, Benítez J, Carrillo JA. Pharmacogenetic testing and therapeutic drug monitoring are complementary tools for optimal individualization of drug therapy. Eur J Clin Pharmacol. 2010; 66(8):755-74.
- Ceppa F, Fontan E, Cremades S, et al. Role of pharmacogenetics in chemotherapy of colorectal cancers. Rev Med Interne. 2007;28(9):594-02.
- Collin LJ, Cronin-Fenton DP, Ahern TP, et al. Cohort Profile: The Predictors of Breast Cancer Recurrence (ProBe CaRE) Premenopausal Breast Cancer Cohort Study in Denmark. BMJ Open. 2018;8(7):e021805.
- Lee SHR, Yang JJ. Pharmacogenomics in acute lymphoblastic leukemia. Best Pract Res Clin Haematol. 2017;30(3):229-36.
- Chantry AS, Quaranta S, Ciccolini J, Lacarelle B. [Clinical application, limits and perspectives of pharmacogenetic and pharmacokinetic analysis of anticancer drugs]. Ann Biol Clin (Paris). 2014;72(5):527-42.
- Cai X, Cao W, Ding H, et al. Analysis of UGT1A1*28 genotype and SN-38 pharmacokinetics for irinotecan-based chemotherapy in patients with advanced colorectal cancer: results from a multicenter, retrospective study in Shanghai. J Cancer Res Clin Oncol. 2013;139(9):1579-89.
- Yang Y, Zhou M, Hu M, et al. UGT1A1*6 and UGT1A1*28 polymorphisms are correlated with irinotecan-induced toxicity: A meta-analysis. Asia Pac J Clin Oncol. 2018 Jun 22:1-11.
- Hall D, Ybazeta G, Destro-Bisol G, Petzl-Erler M.L, et al. Variability at the uridine diphosphate glucuronosyltransferase 1A1 promoter in human populations and primates. Pharmacogenetics. 1999;9(5):591-9.
- Kaniwa N, Kurose K, Jinno H, et al. Racial variability in haplotype frequencies of UGT1A1 and glucuronidation activity of a novel single nucleotide polymorphism 686C> T (P229L) found in an African-American. Drug Metab Dispos. 2005;33(3):458-65.
- Inoue K, Sonobe M, Kawamura Y, et al. Polymorphisms of the UDP-glucuronosyl transferase 1A genes are associated with adverse events in cancer patients receiving irinotecan based chemotherapy. Tohoku J Exp Med. 2013; 229(2):107-14.
- Park SR, Kong SY, Rhee J, et al. Phase II study of a triplet regimen of S-1 combined with irinotecan and oxaliplatin in patients with metastatic gastric cancer: clinical and pharmacogenetic results. Ann Oncol. 2011;22(4):890-6.
- Fukui T, Mitsufuji H, Kubota M, et al. Prevalence of topoisomerase I genetic mutations and UGT1A1 polymorphisms associated with irinotecan in individuals of Asian descent. Oncol Lett. 2011;2(5):923-28.
- Hoskins JM, Marcuello E, Altes A, et al. Irinotecan pharmacogenetics: influence of pharmacodynamic genes. Clin Cancer Res. 2008;14(6):1788-96.
- Fujita K, Sparreboom A. Pharmacogenetics of irinotecan disposition and toxicity: a review. Curr Clin Pharmacol. 2010;5(3):209-17.
- Liu X, Cheng D, Kuang Q, Liu G, Xu W. Association of UGT1A1*28 polymorphisms with irinotecan-induced toxicities in colorectal cancer: a meta-analysis in Caucasians. Pharmacogenomics J. 2014;14(2):120-29.
- IRINOTECAN HYDROCHLORIDE- irinotecan hydrochloride injection [package insert]. Orlando, FL, USA; June 5, 2017. Available from: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=d04f2471-3085-4fc8-a657-bb3918d48e6eu.
- Innocenti F, Schilsky RL, Ramírez J, et al. Dose-Finding and Pharmacokinetic Study to Optimize the Dosing of Irinotecan According to the UGT1A1 Genotype of Patients With Cancer. J Clin Oncol. 2014; 32(22): 2328-34.
- Ichikawa W, Uehara K, Minamimura K, et al. An internally and externally validated nomogramfor predicting the risk of irinotecan-induced severe neutropenia in advanced colorectal cancer patients. Br J Cancer. 2015;112(10):1709-16.
- Li M, Wang Z, Guo J, et al. Clinical significance of UGT1A1 gene polymorphisms on irinotecan-based regimens as the treatment in metastatic colorectal cancer. Onco Targets Ther. 2014;7:1653-61.
- Liu D, Li J, Gao J, Li Y, Yang R, Shen L. Examination of multiple UGT1A and DPYD polymorphisms has limited ability to predict the toxicity and efficacy of metastatic colorectal cancer treated with irinotecan-based chemotherapy: a retrospective analysis. BMC Cancer. 2017;17(1):437.
- Fukuda M, Suetsugu T, Shimada M, et al. Prospective study of the UGT1A1*27 gene polymorphism during irinotecan therapy in patients with lung cancer: results of Lung Oncology Group in Kyusyu (LOGIK1004B). Thorac Cancer. 2016;7(4):467-472.
- Yan L, Wang XF, Wei LM, Nie YL, Liu JY, Zhang LR. Effects of UGT1A1*6, UGT1A1*28, and ABCB1-3435C >T polymorphisms on irinotecan induced toxicity in Chinese cancer patients. Int J Clin Pharmacol Ther. 2016;54(3):193–99.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7-30
- Dowsett M, Cuzick J, Ingle J, et al. Meta-analysis of breast cancer outcomes in adjuvant trials of aromatase inhibitors versus tamoxifen. J Clin Oncol. 2010;28(3):509–18.
- Kalder M, Hadji P. Breast cancer and osteoporosis - management of cancer treatment-induced bone loss in postmenopausal women with breast cancer. Breast Care (Basel). 2014;9(5):312-7.
- de Vries Schultink AHM, Zwart W, Linn SC, Beijnen JH, etal. Effects of pharmacogenetics on the pharmacokinetics and pharmacodynamics of tamoxifen. Clin Pharmacokinet. 2015;54(8):797-10.
- Gaedigk A, Simon SD, Pearce RE, et al. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83(2):234-42.
- Borges S, Desta Z, Jin Y, et al. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J Clin Pharmacol. 2010;50(4): 450-8
- Wigle TJ, Jansen LE, Teft WA, et al. Pharmacogenomics guided-personalization of warfarin and tamoxifen. J Pers Med. 2017;7(4):1-20.
- Goetz MP; Suman VJ, Hoskin TL, et al. CYP2D6 metabolism and patient outcome in the Austrian Breast and Colorectal Cancer Study Group trial (ABCSG) 8. Clin Cancer Res. 2013;19(2):500-07.
- Saladores P, Murdter T, Eccles D et al. Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. Pharmacogenomics J. 2015;15(1):84-94.
- Park IH, Ro J, Park S, et al. Lack of any association between functionally significant CYP2D6 polymorphisms and clinical outcomes in early breast cancer patients receiving adjuvant tamoxifen treatment. Breast Cancer Res Treat. 2012;131(2):455-61.
- Ahern TP, Hertz DL, Damkier P, et al. Cytochrome P-450 2D6 (CYP2D6) genotype and breast cancer recurrence in tamoxifen-treated patients: evaluating the importance of loss of heterozygosity. Am J Epidemiol. 2017;185(2):75-85.
- Dezentjé VO, Opdam FL, Gelderblom H, et al. CYP2D6 genotype- and endoxifen-guided tamoxifen dose escalation increases endoxifen serum concentrations without increasing side effects. Breast Cancer Res Treat. 2015;153(3):583-90
- Binkhorst L, Mathijssen RH, Jager A, et al. Individualization of tamoxifen therapy: much more than just CYP2D6 genotyping. Cancer Treat Rev.2015;41(3):289-99.
- Fox P, Balleine RL, Lee C, et al. DoseEscalation of Tamoxifen in Patients with Low Endoxifen Level: Evidence for Therapeutic Drug Monitoring-The TADE Study. Clin Cancer Res. 2016;22(13):3164-71.
- Gong, I.Y.; Teft,W.A.; Ly, J.; Chen, Y.H.; Alicke, B.; Kim, R.B.; Choo, E.F. Determination of clinically therapeutic endoxifen concentrations based on efficacy from human MCF7 breast cancer xenografts. Breast Cancer Res. Treat. 2013;139(1):61-9.
- Madlensky L, Natarajan L, Tchu S, et al. Tamoxifen metabolite concentrations, CYP2D6 genotype, and breast cancer outcomes. Clinical Pharmacol Ther. 2011;89(5):718-25.
- Martinez de Duenas E, Ochoa Aranda E, Blancas Lopez-Barajas I, et al. Adjusting the dose of tamoxifen in patients with early breast cancer and CYP2D6 poor metabolizer phenotype. Breast. 2014;23(4):400-6.
- Hertz DL, Rae JM. Individualized tamoxifen dose escalation: Confirmation of feasibility, question of utility. Clin Cancer Res. 2016;22(3):3121-23.