Playing with DNA
2 Retd. Reader, Department of Zoology, Cuttack, Odisha, India
Received: 03-Jul-2018 Accepted Date: Aug 17, 2018; Published: 20-Aug-2018
Citation: Mohapatra S, Ghosh S. Playing with DNA. J Clin Gen Genomics. 2018;1(2):14-19.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Today our understanding of “genes” has reached such a level of sophistication and depth that we are no longer studying and altering genes in test tubes but in their native context in human cells. Way back in 1864, in a Moravian Monastery, a priest named “Gregor Johan Mendel” discovered “gene”, though he was not aware of what he had discovered. The word gene was coined a decade later. Later DNA was identified as “gene” the source of genetic information when scientists showed keen interest in deciphering the morphological, structural and functional aspects of genetic material i.e. DNA. In the seventies, two technologies gene sequencing and gene cloning allowed scientists to manipulate genes. Millions of copies of gene hybrid were produced in the test tubes, the hybrids were then inserted into the host cells producing novel gene. In the beginning of this century man was able to read his genomes. The draft sequence of human genome was published in 2001. New tools and techniques were developed to manipulate DNA with more precision and accuracy. Some of them are briefly described in this paper. Scientists are in a cross road, whether they should carry on the program of gene manipulation there by destabilizing the genetic equilibrium which was stabilized in millions of year
Keywords
HGP; CRISPR-Cas9; TALENs; ZFNsIntroduction
No one really has the guts to say it, but if we could make better human beings by knowing how to add genes, why shouldn’t we?
− James Watson, Nobel Laureate
I do not really think our bodies are going to have any secrets left within this century. And so anything that we can manage to think about will probably have a reality.
− David Baltimore, Nobel Laureate
We are living through revolution in human understanding from the dawn of civilization may have been asking questions about why things are the way it is, why we behave as we do, how our bodies work in sickness and in health and why we all seem so similar and yet display such diverse and wonderful individuality. Philosophy, Psychology, Biology, Medicine and Anthropology, even Religion all have attempted to supply answer and not without some success. But very recently, we have been missing a fundamental piece of puzzle with significance to every aspect of human existence.
When the word ‘gene’ was coined, people had very little knowledge about various aspects of gene, little more than fifty years ago, Crick and Watson discovered the structure of the DNA molecule. Soon, it was confirmed that ‘Gene’ is DNA and DNA was crowned as “Master Molecule”. Scientists all over globe started to unravel the secrets of DNA. How it works? Genetic code was discovered in the eighties. Scientists started working on the language of gene.
The whole idea of playing with DNA started in the seventies. In late summer of 1972, at Sicily. Late in the evening, Paul Berg (The American Biochemist who got Nobel Prize in Chemistry, whose work involves gene splicing of recombinant DNA) and a group of students were overlooking the lights of the city when Berg started the conversation on the possibility of counseling two pieces of DNA to create recombinant DNA. This reference actually created tremors of wonder and anxiety among the members present in the conference. The prospects of human genetic engineering i.e. introduction permanently into the human genome gave shock to the scientists present in the conference. They were worried about the future of the future.
From early seventies to 2017, is a long time forty-five years have passed. Now cutting and joining DNA is possible in student’s laboratory. The question what Berg had visualized on that evening what will happen if man plays God?
History
1952: First cloned frog by Briggs and King
1972: The idea of modifying the human genome to treat disease was proposed by Friedmann et al. [1].
1974: First genetic mouse was proposed from viral DNA [2].
1978: July 25; Birth of Louise Brown, first baby born by IVF [3].
1990: Development of PGD (Pre-implantation Genetic Diagnosis) at Hammersmith Hospital in
London and birth of Natalie and Danielle Edwards, twin sisters [4].
1996: July 5; Creation of ‘Dolly’ the sheep by Ian Wilmut and Keith Campbell [5,6].
2002: August 29; Birth of Adam Nash, first child conceived as a ‘savior sibling’ using PGD [7].
2003: Finished Human Genome Sequence published.
2004: Woo-Suk Hwang claims creation of first cloned human embryo.
2005: Successful cloning of a human embryo by a UK team.
DNA: The Master Molecule
In all living organisms, DNA plays a vital role in encoding, storage, replication and propagation of genetic information. It writes the genetic recipes for every living thing on earth.
Each DNA molecule is made up of phosphates and sugars which provide its structural architecture, the nitrogen-rich chemicals known as nucleotides or bases, which encode genetic information. The bases come in four varieties: Adenine (A), Cytosine (C), Guanine (G) and Thymine (T). Together these provide the letters in which the genetic code is written.
The bases can be further subdivided into two classes: Adenine and Guanine are larger structures called purines; Cytosine and Thymine are smaller pyrimidines. Each purine has complementary pyrimidine to which it will bind-A is complementary to T and C is complementary to G. It is software of life, containing the information needed to build and run a body.
The DNA molecule is composed of two linked chains of bases. Each base is joined to its partner by hydrogen bond A to T and C to G and in the other end held by a sugar and phosphate backbone. Two DNA strands twist around each other in a double helix. Each strand is the mirror image of the other, where one has an A, its partner will always have a T and vice versa. If the first strand reads ACGTTACCGGTC, the other will read TGCAATGGCCAG.
The Human Genome Project
By deciphering the sequence of whole human genome and a key model in an integrated manner, HGP made huge transformation in molecular and biological research. It is an international effort to clone and sequence the entire human genome.
It is the first internationally coordinated effort in the history of biological research. The project was aimed to find out the sequence of entire human genome comprising about 3 billion base pairs.
HGP (Human Genome Project) was a first in its kind of collaborative project and remained world’s largest project in collaboration having its major contributors like NHGRI at National Institute of Health and Department of Energy (DOE), USA. Apart from these there were several international contributors; universities and laboratories from Japan, China, UK, Germany and France. The first funding was released by NHGRI in 1988. However, the discussion about the project was started as early in 1984.
The Story Of DNA Manipulation
In late 1974, the invention of recombinant DNA technology a genemodified SV40 virus was used to infect early mouse embryonic cell. The plan was audacious. The virus infected embryonic cells were mixed with the normal embryo to create a composite of cell, an embryological ‘chimera’. These composite embryos were implanted into mice. They differentiated into all sorts of cells-blood, brain, muscle, liver and even sperm and egg cells. The experimental modification the scientists were doing in petri-dishes becomes the genetic modification of an organism in a womb. It was a transition between lab and life.
The experimental ease permitted by embryonic stem cells also surmounted a major problem. When viruses were used to deliver a gene into cell, it is virtually impossible to control where the gene inserted into the genome. At three billion base pairs of DNA the human genome is about fifty thousand or a hundred through times the size of most viral genome. A viral gene drops like a small coin dropped into Atlantic from an aeroplane. There is no way to predict where it will laud. Virtually all viruses capable of gene integration such as HIV or SV40 generally lack their genes randomly into same spot in human genome. For gene therapy, this random integration is an infernal nuisance. The viral gene may fall into an area that is not desirable. Worst the integration, might disrupt an essential gene or active cancer-causing gene resulting in potential disasters.
Scientists learned to make genetic changes in targeted position in the genome using the embryonic stem cells. This technology had a farreaching implication. In the natural world, the only means to achieve a directional intellectual change in a gene is through random mutation and natural selection. Suppose an animal is exposed to X-rays, a genetic alteration take place, but this change is random. Natural selection must choose the mutation that confers the best fitness to the organism thereby allows that mutation to become increasingly common in the gene pool.
In nature, the engine that drives genetic alteration has no one in its driver’s seat. The watch maker of evolution as Richard Dawkins reminds us in blind.
Using ES cells however a scientist would designedly manipulate just about any chosen cistron and incorporate that genetic modification for good into the ordering of an animal. Transgenic animals are seen in different laboratories of world by early 1990s. Hundreds of strains of transgenic mice had been created. One mouse was prepared with a Jellyfish gene inserted into its genome that allowed it to glow in the dark under blue lamp.
In 1988, a two-year old girl named Ashauti De Silva (Ashi) from Ohio began to develop peculiar symptoms. Ashi’s illness and symptoms were markedly abnormal: bizarre pneumonia and infection which seems to persist wounds that would not heal. The culprit of the syndrome was ADA gene-short for Adenosine Deaminase encodes an enzyme that converts Adenosine a natural chemical produced by the body into a harmless product called Inosine.
In the absence of ADA gene, the detoxification reaction fails to occur, and the body gets clogged with toxic-byproducts of Adenosine metabolism. ADA deficiency is a part of a larger group of notorious illness called Severe Combined Immunodeficiency (SCID).
Could gene therapy correct ADA deficiency? After all, only one gene needed to be corrected and the gene had already been identified and isolated. A vehicle or vector designed to deliver genes into human cells had also been identified. In 1980, at National Institute of Health, a team of gene therapist led by William French Anderson and Michael Blaese decided to deliver ADA gene into children with ADA deficiency. The first human trial was carried on Ashi now four years old. The first attempt of Dr. Anderson on human was partly successful. The parents were convinced but Ashi continued to be on medication. The most ambitious gene trial attempted in human was measured in the frequency of running nose and some for the parameters. Scientists are still intellect regarding the success of the medical aid on human.
New Tools In DNA Editing
From 1988 to 2014, there was a big gap; meanwhile various types of restriction enzyme were discovered. In the recent decades, human ordination engineering has been one amongst the most important fascinating analytical subjects; primarily as a result of it raises new prospects for customized drugs and biotechnologies. With the event of designed nucleases such as ZFNs, TALENs and a lot of recently CRISPR, the field of human ordination edition has evolved terribly chop-chop.
Engineered nucleases have modified the approach that we tend to use edited genetic data. It is currently potential to focus on specific genetic changes in an exceedingly selected locus, to either insert or edit polymer.
A designed virus containing the custom designed nuclease and the emended nucleotide DNA sequence is unit transfected enzyme that cleaves a selected website of chromosomal DNA to induce Double Stranded Break (DSB) victimization the cell's DNA repair machinery.
ZFNs
It includes a robust category of tools that are unit redefining the boundaries of scientific research. It’s composed of programmable, sequence specific DNA-binding modules coupled to a non-specific deoxyribonucleic acid cleavage domain. It allows a broad range of genetic modifications by causing DNA Double Stranded Breaks that stimulate fallible Non-Homologous End Joining (NHEJ) or Homology-Directed Repair (HDR) at specific genomic locations [8,9].
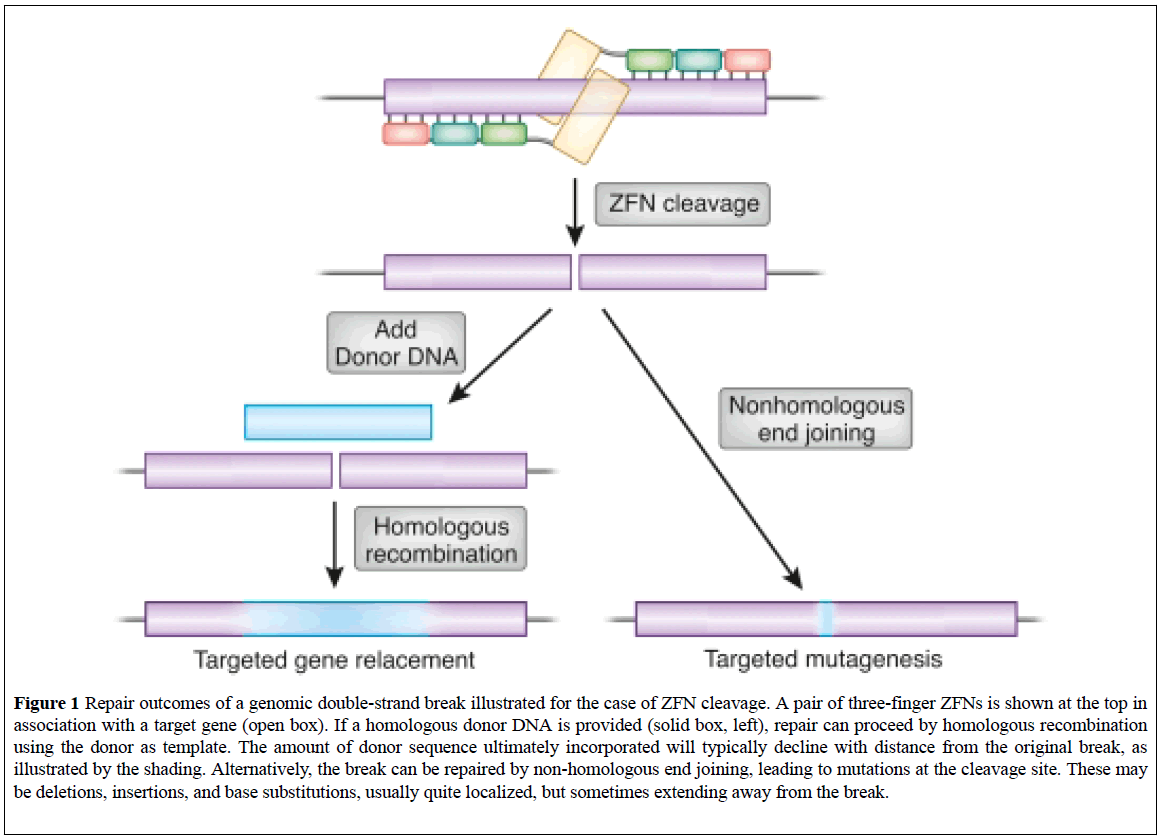
These are targetable DNA cleavage reagents that have been adopted as gene targeting tools. ZFN-induced DSBs are subjected to cellular DNA repair processes that lead to both targeted mutagenesis and targeted gene replacement at remarkably high frequencies. ZFN-mediated genome altering takes place within the nucleus when a pair of ZFNs targeting the user's gene of interest (GOI) is delivered transiently into a cell line, either by transfection or electroporation (Figure 1) [10,11].
Figure 1: Repair outcomes of a genomic double-strand break illustrated for the case of ZFN cleavage. A pair of three-finger ZFNs is shown at the top in association with a target gene (open box). If a homologous donor DNA is provided (solid box, left), repair can proceed by homologous recombination using the donor as template. The amount of donor sequence ultimately incorporated will typically decline with distance from the original break, as illustrated by the shading. Alternatively, the break can be repaired by non-homologous end joining, leading to mutations at the cleavage site. These may be deletions, insertions, and base substitutions, usually quite localized, but sometimes extending away from the break.
TALENs
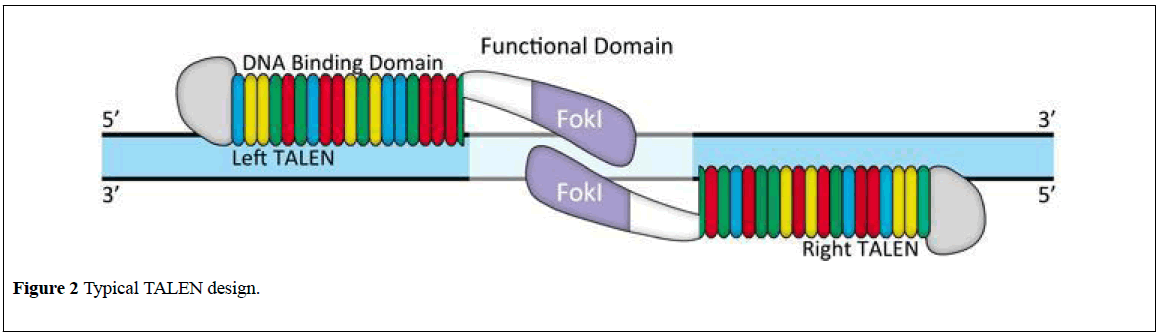
TALENs refer to Transcription Activator-Like Effecter Nucleases. TALEN is comprised of a pair of DNA binding proteins fused to the FokI nuclease (Figure 2) [12], TALENs comprise a non-specific DNA-cleaving enzyme united to DNA-binding domain which will be simply designed in order that they can target basically any sequence the capability to quickly and with efficiency altered genes victimization. TALENs guarantees to own profound impacts on scientific research and to yield potential therapeutic ways for genetic diseases.
TALENs have recently emerged as a revolutionary genome editing tool in many varieties of organisms and cells. The site-specific chromosomal DSBs introduced by TALENs significantly increase the efficiency of genomic modification.
CRISPR-cas9
CRISPR-Clustered Regularly Interspaced Short Palindromic Repeats are segments of prokaryotic DNA containing short repetition of base sequences. Every repetition is followed by short segments of spacer deoxyribonucleic acid [13].
It's an individual technology that permits geneticists and medical researchers to edit components of the order by removing, adding or neutering sections of the DNA sequence.
CRISPR-cas9 technology is relatively simple to style because it solely needs ever-changing the sgRNA sequence to target the desired locus and therefore will be engineered applying molecular biological research and synthesis procedures [14].
Presently the best, precise and most versatile technique of genetic manipulation is CRISPR that is inflicting a buzz within the science world. This system presently stands out because the quickest, least expensive and most reliable system for altering material genes.
Who Invented Crispr-Cas9?
In 1987, at Osaka University, Yoshizumi Ishino was finding out the E. coli gene 'iap'. Throughout their sequencing efforts, they found this odd twenty nine nucleotide repeat sequence with a thirty two nucleotide spacing.
In 2000, a team of Francisco Mojica and Guadalupe Juez' at Alicante University did a fast comparative analysis of prokaryote genomes and known a series of Short Regularly Spaced Repeats (SRSRs) that were common among multiple species.
In 2002, Rudd Jansen of Utrecht University once more found a 21-37 bps repeat and citing the Ishino and Mojica work, recognized that these interspaced short sequence repeats have a definite spacing that varies by organisms. Notably, they found that CRISPRs were unique to certain prokaryotes and not viruses and eukaryotes.
They found a typical sequence, at the ends GTT/AAC of the ends. Upstream of the CRISPR loci, there was a long homologous sequence while not associate open reading frame indicative of a preserved American state polymer phase. Nearby, there were able to establish homologous genes which might become cas1 to cas4. Working with Mojica, they came up with the name CRISPR [15]. It was then realized that Ishino happened to discover the first CRISPR loci.
A chemist Dr. Jennifer Doudna at the University of California, Berkeley, helped create one in all the foremost monumental discoveries in biology: a comparatively easy way to alter DNA of any organism, even as a human will edit a word during a document. The questionable CRISPR-cas9 ordination piece of writing technique is already wide employed in laboratory studies and scientists hope it's going to facilitate rewriting blemished genes in folks, gap tremendous new prospects for treating and natural action diseases in the future.
How Was Crispr-Cas9 Developed?
Some microorganisms have an analogous, constitutional sequence written material system to the CRISPR-cas9 system that they use to reply to invade pathogens like viruses, very similar to associate in nursing system.
Victimization CRISPR, the microorganisms snip out elements of the virus deoxyribonucleic acid and keep a small amount of it behind to assist them acknowledge and defend against the virus next time it attacks. Scientists tailored this method in order that it can be utilized in different cells from animals together with mice and humans.
Scientists custom-made this technique, so it may be utilized in alternative cells from animals together with mice and humans.
Mechanism
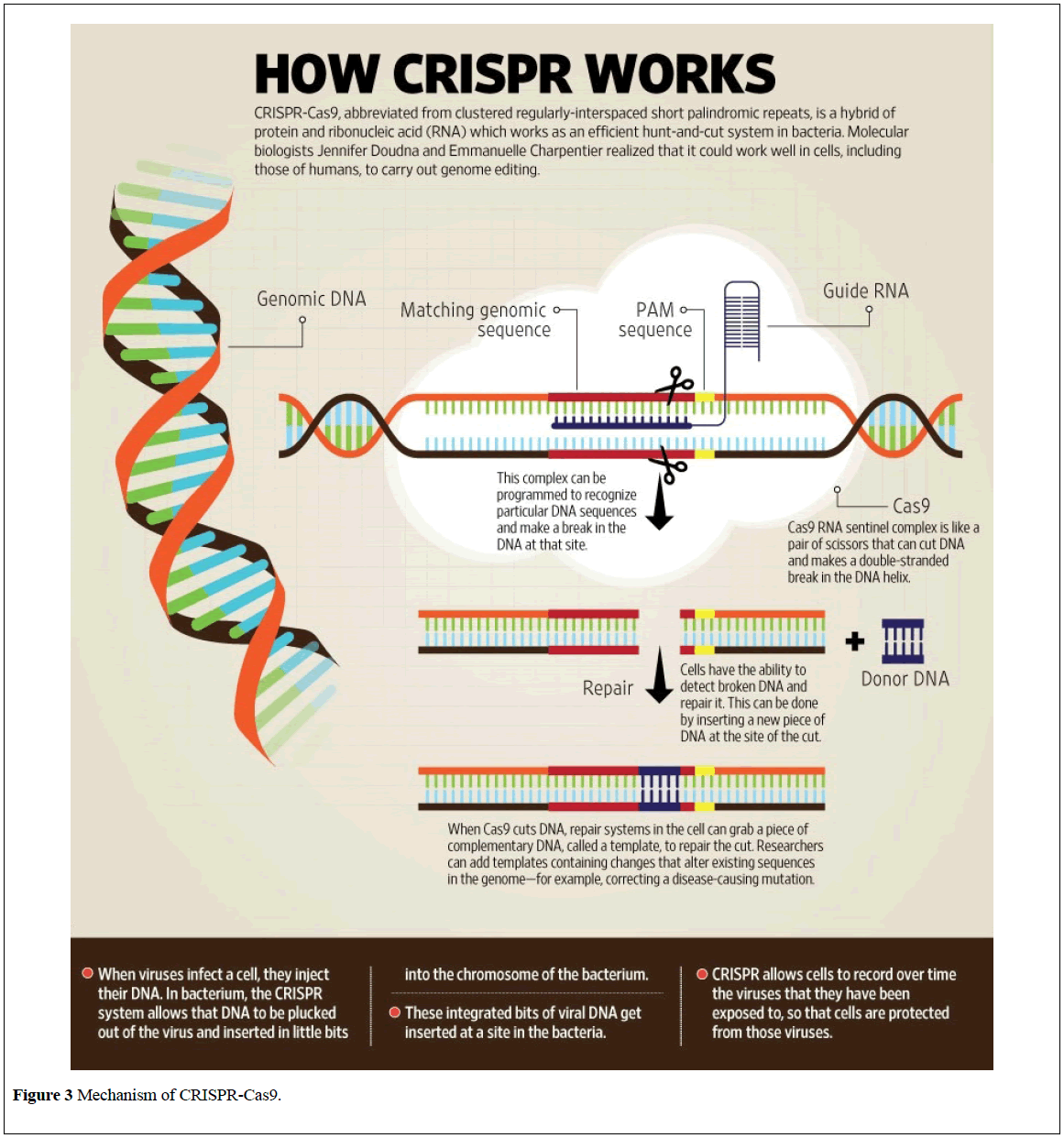
This possibility of change our fate has become more realistic with the development of CRISPR-Cas9 [16]. It is the use of an enzyme to remove pieces of genetic code and replace them with a different sequence to alter characteristics of a human being (Figure 3) [17].
The key step in editing the genome of an organism is selective targeting of a specific sequence of DNA. The CRISPR-cas9 system consists of 2 key molecules that introduce a mutation into the DNA. These are:
AN ENZYME cas9 – It functions as a cellular surgical knife which will cut the 2 strands of deoxyribonucleic acid at a particular location within the ordination in order that bits of deoxyribonucleic acid can then be removed.
A PIECE OF RNA – This is referred to as guide polymer (g-RNA) that leads the surgical knife to the precise nucleotides. This consists of a little piece of pre-designed polymer sequence that is regarding twenty bases long set among extended polymer scaffold. The scaffold half binds to polymer and therefore the pre-designed sequence guides cas9 to the proper a part of the ordering. This makes positive that the cas9 accelerator cuts at the proper purpose in the ordering.
The guide RNA is meant to seek out and bind to a selected sequence within the polymer. The guide RNA has RNA bases that area unit complementary to those of the target polymer sequence within the ordering. This suggests that the guide RNA can solely bind to the target sequence and no alternative regions of the ordering [16].
The cas9 follows the guide RNA to an equivalent location within the deoxyribonucleic acid sequence and makes a cut across each strand of the deoxyribonucleic acid. At this stage the cell acknowledges that the deoxyribonucleic acid is broken and tries to repair it. Scientists will use the deoxyribonucleic acid repair machinery to introduce changes to at least one or a lot of genes within the order of a cell of interest.
Applications for Crispr-Cas9 Technology
Treating disease
CRISPR-cas9 has a lot of potential as a tool for treating a range of medical conditions that have a genetic component, as well as cancer, viral hepatitis or maybe high sterol.
Many of the planned applications involve redaction the genomes of corporal cells however there have been tons of interest in and discussion concerning the potential to edit germ line.
Genetic diseases most amenable for CRISPR-Cas9 writing area unit those within which one factor must be targeted [17].
The distinctive multiplexing ability of CRISPR-Cas9 might take synchronous targeting risk in close to future [18].
CRISPR-Cas9 will be accustomed disrupt the dominant gene by NHEJ for diseases that result from the merchandise of infective cistron merchandise.
Altering ecology
The unfold of vector-borne sicknesses like protozoal infection may well be reduced by introducing sickness resistant genes into wild insect populations.
Transforming food
CRISPR can be wont to develop drought resistant or otherwise hardier crops.
Editing humans?
CRISPR has the potential to edit human but should we do that?
Future Aspects of Crispr-Cas9 Technology
It is likely to be many years before CRISPR-Cas9 is used routinely in humans.
Much research is still focusing on its use in animal models or isolated human cells with the aim to eventually use the technology to routinely treat diseases in humans.
There is a lot of work focusing on eliminating ‘off target' effects, wherever the CRISPR-Cas9 system acts as a distinct factor to the one that was supposed to be altered.
No scientific discovery of the past century holds a lot of promise or raises more worrisome moral queries. Most provokingly if, CRISPR were accustomed edit embryos of a person's germ line cells that contain genetic material which will be hereditary by subsequent generation either to correct genetic flow or to boost a desired attribute, the modification would then pass to that person's youngsters and youngsters in sempiternity. The total implications of changes that profound area unit tough, if not possible to foresee. This could be an exceptional technology with several uses; however, if you're getting to do something as fateful as redaction the germ line, you'd higher be able to tell Pine Tree State there's a robust reason to try to it − said WHO served as a frontrunner of HGP (currently Director MIT).
CRISPR-Cas9 is sort of a double-edged blade. Last year, in a very study printed within the PNAS (Proceedings of National Academy of Sciences) CRISPR was accustomed engineer a version of mosquitoes that creates them incapable of spreading the protozoal infection genus Anopheles parasite. A little package of deoxyribonucleic acid was additional to the dipterous insect the modification within the sequence may forestall the deadly parasite from being transmitted by the mosquitoes.
The possibility of adjusting the incorrect cistron and rising the lifetime of millions could be a commendable achievement. Replacement polygenic disease cistron or Alzimer's cistron square measure within the pipe line, equally genetically built crops, resisting herbicides and insects began to rework a lot of the world's agricultural landscape. Scientists in Japan have used CRISPR to increase the lifetime of tomatoes by deleting all 3 copies of 1 wheat cistron. A replacement strain has been created that may resist the mildew unwellness. Shortly doctors are also able to use CRISPR to treat some specific cancer disease directly.
Conclusion
The big question should we play God by changing the natural system. What is natural is neither good nor bad. Francis Collins Director NIH writes “Ethical issues presented by altering the germ line in a way that affects the next generation without their consents”. How can you make decisions for future?
The next big question altering gene will destabilize ‘gene pool’. We are surviving for a stable ‘gene pool’. It took millions of years to achieve this stability. Disturbing the ‘gene pool’ won’t disastrous???
REFERENCES
- Friedmann T, Roblin R. Gene therapy for human genetic disease? Science 1972;174:949-55.
- Jaenisch RM, Mintz B. Simian Virus 40 DNA Sequences in DNA of Healthy Adult Mice Derived from Pre-implantation Blastocysts injected with viral DNA. Proc Natl Acad Sci. 1974;71:1250-4.
- Hutchinson, Martin. “I helped deliver Louise”. 2003.
- Henderson M. 50 Genetics Ideas You really need to know, Quercus Editions Ltd. 2008, Chapter 41-Designer Babies.
- McLaren A. “Cloning: pathways to a pluripotent future”. Science 2000;288:1775-80.
- Wilmut I, Schnieke AE, McWhir J, et al. “Viable offspring derived from fetal and adult mammalian cells”. Nature 1971;385:810-3.
- Horsey K. “US ‘savior sibling’ spark debate”. Progress Educational Trust.
- Christian M, Cermak T, Doyle EL, et al. Targeting DNA double strand breaks with TAL effector nucleases, Genetics 2010;186:757-61.
- Pattanayak V, Ramirez CL, Joung JK, et al. Revealing of target cleavage specificities of Zinc-Finger Nucleases by In Vitro Selection, Nature methods 2011;8:765-70.
- Seokjoong Kim, Jin-Soo Kim. Targeted Genome Engineering via Zinc Finger Nuclease 2011;5:9-17.
- Carroll D. Genome Engineering with Zinc-Finger Nucleases. Genetics 2011;188:773-82.
- Davis Ed. Genome Editing: Which Should I Choose, TALEN or CRISPR? 2018.
- Jinek M, Chylinski K, Fonfara I, et al. A programmable deal- RNA guided DNA endonuclease in adaptive bacterial immunity, Science 2012;337:816-21.
- Mali P, Yang L, Esvelt KM, et al. RNA guided human genome engineering via Cas9,Science 2013;339:823-26.
- Carr S. Looking-beyond-the-debate-of-who-owns-CRISPR-gene-editing-technology. 2017.
- Galton F. EUGENICS: its definition, scope, and aims.1904.
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR-Cas systems, science 2013;339:819-23.
- Ye L, Wang J, Beyer AI, et al. Seamless modification of wild- type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection, Proceedings of the National Academy of sciences of the United States of America, 2014;111:9591-6.