Prevalence of microbial contamination of mobile cell phones in general population of Delhi, India
2 Deaprtment of Microbiology, Shriram Institute for Industrial Research, 19, University Road, Delhi, India, Email: dushyant.singh@gmail.com
Received: 12-Oct-2018 Accepted Date: Nov 04, 2018; Published: 14-Nov-2018
Citation: Chauhan A, Garg S, Ranjan AA, et al. Prevalence of microbial contamination of mobile cell phones in general population of Delhi, India. J Exp Clin Microbiol 2018;1(1):12-15.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Keywords
Mobile cell phones; Microbial contamination; General population; Microbial infectionsIntroduction
Mobile phone users in India have increased gradually that makes about 5 billion populations in the year 2017. Mobile phones are the hand held devices that are a major part of communication system. It enables people to make contact to each other. Apart from its function as means of communication, they have evolved due to human intelligence after the collaboration of various soft was within a single mobile device. They are used by all types of people irrespective of their social or occupational background. From a rickshaw puller to the owner of a high-tech company, everyone uses a mobile because it is just like any other need that have helped people connected to communicate. This device keeps the population updated with the surroundings and had maintained the work pace of the cooperate world. Mobile phone usage all over the world has increased tremendously. New applications and technologies have troubled the lives of human. They offer as a source of contamination. It is necessary to aware public regarding the prevention of infection and highlighting their duty to control infections [1]. Our phone remains in close contact with our body, majorly with our hands, mouth, ears and face. Apart from this, it is very common practice of carrying the phone to washroom. Also, people have habit of operating phone while consuming meals permitting the direct transfer of microorganisms from the mobiles to our gut.
Mobile phones are absolutely essential devices and majorly operated in the areas of excessive microbial load. Firm actions must be incorporated in our daily activities like proper washing of hands before and after meals, cleaning of phones with disinfectant [2] .There is no such known practice to clean the mobile phones that result in its contamination by the microbes. We can view the dust by our naked eyes that get deposited on the mobiles surface but unable to see the microbes that would be flourishing on phone. Akineymi, et al. determined the contamination of microbes on the phone of food vendors and marketers which was found to be very high that could be related to the poor sanitation maintained by people in their surroundings or the lack of knowledge.
These days, mobile phones have proved to be the most important hand held device for professional as well as personal work. It is used daily everywhere by households, workers, healthcare staffs, etc as communication medium. No strict rules have been followed to avoid mobile phones in ICU where contamination risk is higher. Reasonable prices have turned the public dependency on mobile phones exponentially. Mobile phones being in touch with our body and different objects are creating frequent health issues. Sanitizing cell phone is itself a cumbersome task. In the study conducted, 77.11% of phones showcased microbial contamination [3].
The important preventive measure is maintaining the hygienic conditions of hands to avoid cross contamination and infections. Hand comes in contact with our phones very frequently therefore serving as an infectious source. In a study done in hospitals, 74% of the mobile phones were found contaminated [4]. Development of prevention plans like regular cleaning of mobile phones with the disinfectants is able to minimize the disease vulnerability. Using antimicrobial methods to avoid the bacterial contamination is very easy by practicing appropriate techniques to clean the phone. Concern can be shown in the production of mobile phones with some safe material that would inhibit the bacterial growth [5]. Due to the frequent usage of mobile phones, heat generated by the phone creates an ideal temperature that supports the growth of bacteria. The possibility of getting an infection due to mobile phone is high as they as kept in very close proximity or touch with our face, hands and mouth providing a route to bacteria to enter the body.
Till date, there is no report on the degree of microbial contamination including common human pathogen of general population of Delhi such as housewives, working people, shopkeepers, street vendors and health care staff. The present study was carried out to know the record of total microbial count (bacterial, Yeast & Mould), detection, isolation and identification of commonly pathogenic microorganisms like E. coli, Pseudomonas, S. aureus and bacillus from mobile phones of different people collected from various locations.
Materials and Methods
Sample collection
25 samples of different people from random locations were collected by a sterile swab method that is ISO 18593:2004. A collection kit was made that included IPA, cotton, gloves, swabs having 9 ml of 0.1% tryptone water and paraffin tape for sealing the collected swabs (Figure 1). The samples were collected in duplicate and taken to the laboratory in refrigerating conditions for performing the tests.
Biological media, chemicals and reagents
Biological media such as Total Plate count, Nutrient broth, Chloramphenicol Yeast Glucose Agar, Baird parker agar, Ceramide Agar, Eosin Methylene Blue agar, MacConkey agar, Skimmed milk agar, Huge leifson agar, nitrate broth, gelatin medium, Starch agar, Kovacs reagent. All the media and reagent were procured form Hi-Media Laboratory, Mumbai, India. Procured dehydrated media were used as per the instructions written on the box and growth promotion test of each media carried out before evaluation of samples. Sodium chloride, sodium thiosulphate and other chemicals were of analytical grade. Rabbit plasma, anti-sera O, H, Vi, Gram-stain kit and other reagents were procured from Difco Laboratories.
Quantitative estimation of microorganisms
Enumeration of total bacterial count: 1 ml of the sample from the swab to sterile Petri dish is transferred and further the initial dilution was prepared by adding 1 ml of sample in 9 ml diluent (1:10 dilution) of which 1 ml was transferred to 9 ml diluent making dilution 10-2 and repeating the dilution up to 10-5. Each dilution was transferred to respective Petri dishes followed by pouring about 15 ml Plate Count Agar (PCA HiMedia) which was allowed to solidify and incubate at 37°C for 24h [6].
Enumeration of total yeast & mould count: 1 ml of the sample from the swab to sterile Petri dish is transferred and further the initial dilution was prepared by adding 1 ml of sample in 9 ml diluent (1:10 dilution) of which 1 ml was transferred to 9 ml diluent making dilution 10-2 and repeating the dilution up to 10-5. Each dilution was transferred to respective Petri dishes followed by pouring about 15 ml Chloramphenicol Yeast Glucose Agar (CYGA HiMedia) which was allowed to solidify and incubate at 25°C for 5 days.
Qualitative estimation of microorganisms
Isolation and identification of Bacillus cereus: Biological media such as Total Plate count, Nutrient broth, Chloramphenicol Yeast Glucose Agar, Baird parker agar, Ceramide Agar, Eosin Methylene Blue agar, MacConkey agar, Skimmed milk agar, Huge leifson agar, nitrate broth, gelatin medium, Starch agar, Kovacs reagent. All the media and reagent were procured form Hi-Media Laboratory, Mumbai, India. Procured dehydrated media were used as per the instructions written on the box and growth promotion test of each media carried out before evaluation of samples. Sodium chloride, sodium thiosulphate and other chemicals were of analytical grade. Rabbit plasma, anti-sera O, H, Vi, Gram-stain kit and other reagents were procured from Difco Laboratories.
Quantitative estimation of microorganisms
Enumeration of total bacterial count: 1 ml of the sample from the swab to sterile Petri dish is transferred and further the initial dilution was prepared by adding 1 ml of sample in 9 ml diluent (1:10 dilution) of which 1 ml was transferred to 9 ml diluent making dilution 10-2 and repeating the dilution up to 10-5. Each dilution was transferred to respective Petri dishes followed by pouring about 15 ml Plate Count Agar (PCA HiMedia) which was allowed to solidify and incubate at 37°C for 24 h [6].
Enumeration of total yeast & mould count: 1 ml of the sample from the swab to sterile Petri dish is transferred and further the initial dilution was prepared by adding 1 ml of sample in 9 ml diluent (1:10 dilution) of which 1 ml was transferred to 9 ml diluent making dilution 10-2 and repeating the dilution up to 10-5. Each dilution was transferred to respective Petri dishes followed by pouring about 15 ml Chloramphenicol Yeast Glucose Agar (CYGA HiMedia) which was allowed to solidify and incubate at 25°C for 5 days.
Qualitative estimation of microorganisms
Isolation and identification of Bacillus cereus: 4 ml of the sample was inoculated in 50ml of Nutrient Broth and incubated for 24h at 37°C. A loopful inoculum from Nutrient Broth was further streaked onto MacConkey Agar (MCA HiMedia) plate [7]. On observation of pink colonies on plate, tests were performed for confirmation of isolate. These colonies were streaked on NA slant and incubated for 24 h at 37°C which are then streaked on Sheep Blood Agar plate. After the plates have been incubated for 24 h at 30°C, they are observed for the clear zone around colonies for detecting the haemolytic activity of the isolate.
Isolation and identification of Staphylococcus aureus: 4 ml of the sample was inoculated in 50 ml of Nutrient Broth and incubated for 24 h at 37°C. A loopful inoculum from Nutrient Broth was further streaked onto Baird Parker Agar (BPA HiMedia) plate. On observation of shiny black colonies on plate. These colonies were tested for their conformation as S. aureus by streaking the colonies on NA slant and incubating it for 24 h at 37°C. A loop full of culture taken from NA slant was inoculated in test tube containing 50 μL Brain Heart Infusion Broth and 500 μL Rabbit plasma which was then observed periodically for the coagulation at an interval of 1 h [8].
Isolation and identification of E. coli: 4 ml of the sample was inoculated in 50ml of Nutrient Broth and incubated for 24 h at 37°C. A loop full of inoculum from Nutrient Broth was further streaked onto MacConkey Agar (MCA HiMedia) and Eosine Methylene Blue Agar (EMB HiMedia) agar plate. On observation of pink colonies on MCA plate & Green metallic sheen colonies on EMB plate and to confirm the same, biochemical tests were performed (Chauhan and Goyal, 2013). Indole ring test is very common to detect E. coli. Colonies from the EMB plate were streaked on NA slant and incubated for 24 h at 37°C. This slant is used to set the density of culture obtained which is inoculated in typtone broth and incubated for 24 h at 37°C. Now, upon addition of Kovac’s reagent a cherry red ring would appear if the culture was positive for E. coli. [8].
Isolation and identification of Pseudomonas aeruginosa: 4 ml of the sample was inoculated in 50ml of Nutrient Broth and incubated for 24 h at 37°C. A loop full of inoculum from Nutrient Broth was further streaked onto Cetrimide Agar (CA HiMedia) plate. On observation of green fluorescent colonies on plate, biochemical tests were performed for confirmation of isolate. Colonies are taken from CA plate and streaked on NA slant which are incubated for 24 h at 37°C. This slant is used to proceed for biochemical tests that includes streaking on Skim Milk Agar and Starch Agar plates that have to be incubated at different temperatures. Also, a loop full of culture is inoculated in 10 ml Hugh Leifson Medium, 10 ml Gelatin media and 5 ml Nitrate Broth. Oxidase and catalase tests are an important part of these biochemical test confirmation. Oxidase disc (HiMedia) were used to test for oxidase activity and inoculating a loopful of culture from NA slant to a slide with one or two drops of hydrogen peroxide gives the results for production of catalase enzyme by the isolate.
Morphological Identification of isolates: Gram staining was performed for all the isolates to characterize and classify the bacteria. A small amount of culture from the respective NA slants of the isolates were transferred to clean slide with a drop of 0.85% saline. A thin smear is made by rotating the loop in circular motion. This smear was air dried and heat fixed to avoid the smear from being washed off. Cover the smear by pouring crystal violet stain for 30 seconds, after which slide is tilted to pour off the stain and washed with distilled water. Now the slide is covered with iodine solution for 30 seconds and then washed off with distilled water followed by decolorizer. Before applying the counter stain, slide is washed with distilled water and then covered with safranin (counter stain) for 60 seconds. A last wash is given with distilled water and slide is kept for air drying. Now it can be viewed under microscope at 100X after putting a drop of emulsion oil on the area of slide to be observed [9,10].
Results
The way it had changed our life style is commendable and this is one of the reasons for it to become very common device among population. Maximum people have access to the cell phones because of its easy handling and availability in various prices according to user feasibility. We conducted a study to test mobile phones for the microbial contamination. Mobile phones can have multiple users. This creates a frightening situation as it allows the cell phone to harbor the pathogens for different people and also exposing the individual to new micro flora. Therefore this study helps to highlight the extent to which the mobile phones are affected and prove them as a carrier for infections and diseases. All the samples collected exhibit presence of greater number of microorganisms. These microorganisms are known for causing infections in different body parts and can cause serious health issues.
Quantitative estimation of microorganisms
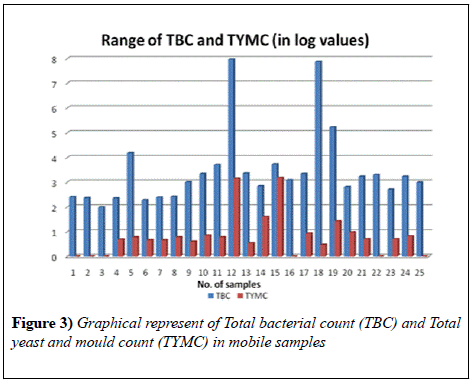
The samples collected for testing the presence of pathogens on mobile phones revealed a high contamination. Results revealed that the Total bacterial count in all the 25 samples ranged from 97 to 9.3 × 107 cfu swab-1 and the Total Yeast and Mould count in 19 samples ranged from 3 to 1.5 × 103 cfu swab-1. Maximum bacterial load was found in was found in the cell phone of a shopkeeper and minimum in that of a Housewife. Also, the maximum load of yeast and mould was observed in the mobile phone of a shopkeeper whereas minimum in housewives.
Qualitative estimation of microorganisms
Isolation and identification of Bacillus cereus: The sample M-2, M-4, M-5, M-6, M-7, M-8, M-9, M-10, M-12, M-13, M-15, M-16, M-17, M-18, M-19, M-21, M-22, M-23, M-24 and M-25 showed the presence of B. cereus which was first isolated on the selective media MYP; the characteristic pink colonies were further identified by biochemical tests. The isolates were compared with the reference positive control B. cereus ATCC 11778, then confirmed as B. cereus. Gram positive rod structures were observed after gram staining the culture. The haemolysis test performed on Sheep Blood Agar was positive for the above mentioned samples as a very clear zone could be seen due to the beta haemolysis around the colonies that grew on Blood Agar.
Isolation and identification of Staphylococcus aureus: The samples M-1, M-3, M-4, M-5, M-6, M-7, M-8, M-9, M-10, M-11, M-12, M-13, M-14, M-15, M-16, M-17, M-18, M-19, M-20, M-22, M-23, M-24 and M-25 showed the presence of S. aureus which was first isolated on the selective media BPA; the shiny black characteristic colonies were further identified by biochemical tests.The isolates were compared with the reference positive control S. aureus ATCC 6538, then confirmed as S. aureus. Gram positive cocci structures were visible upon Gram staining. Gel formation was seen in the coagulase test performed that gives a positive result for the presence of S. aureus.
Isolation and identification of E. coli: The sample M-17 showed the presence of E. coli which was first isolated on the selective media EMB and MCA; the characteristic green metallic sheen colonies were further identified by biochemical tests. The isolates were compared with the reference positive control E. coli ATCC 11105, then confirmed as E. coli. Isolate showed gram negative rods upon gram staining. A cherry red color ring formation occurred on performing Indole test which indicates the presence of E. coli. Also, it was found positive for the motile and positive for the test of Lactose, Glucose, Sucrose and Sorbitol fermentation. The citrate utilization test result was negative.
Isolation and identification of Pseudomonas aeruginosa: The sample M-14 and M-15 showed the presence of P. aeruginosa which was first isolated on the selective media CA the characteristic light green colonies were further identified by biochemical tests. The isolates were compared with the reference positive control Pseaeruginosa ATCC 9027, then confirmed as P. aeruginosa. Gram negative rod structures of P. aeruginosa were visible on gram staining. Positive results were obtained in catalase test confirming the presence of P. aeruginosa due to the formation of bubbles on adding the culture to hydrogen peroxide. Test for oxidase was also positive as the oxidase disc turned from white colour to blue. Starch hydrolysis was absent for these cultures that is a negative result on Starch Agar plate because no clear zone was seen around the colonies. Isolate culture that was stabbed in Hugh Leifson Medium gave yellow colour on incubation for 48 h at 37°C showing positive result for P. aeruginosa. Gelatin was liquified thus giving a positive result for Gelatin test. Nitrate presence was found in these two samples when 0.5 ml each of Sulphanilic acid and α- naphthylamine solution was added to inoculated nitrate broth kept for 24 h. Development of red colour showed the positive result. Growth was observed on Skim Milk Agar with a clear zone around the colonies in plates incubated at 37°C and 44°C which describes the hydrolytic activity of the bacteria. This growth was absent on the plates incubated at 4°C as such low temperature don’t support the survival of isolate.
Discussion
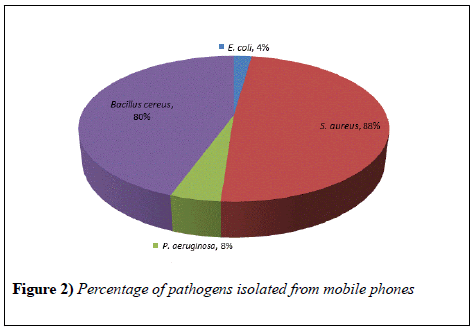
The study showed that all the mobile phones were found to be contaminated with pathogens. Major isolate from the samples was S. auerus, identified in 23 (88%) mobile phones. This is a gram negative bacteria having round shape present in the nose, respiratory tract and on the skin. It has potential to cause different superficial pyogenic infections of dermis and can cause minor infections as well as serious diseases like meningitis, abscesses, etc. Bacillus cereus was also isolated in good number that is in 21 (80%) mobile phones (Figure 2). They are capable of surviving in various stressful conditions and known for the production of toxins which could result in diarrhea, nausea and vomiting. Bacillus show quick multiplication at room temperature. E. coli and P. aeruginosa were present in 2 (8%) and 1 (4%) mobile phone respectively. E. coli bacteria are usually present in environment, food and human and animal intestine. It is responsible for different illnesses like diarrhea, urinary tract infection, pneumonia, etc. P. aeruginosa are capable of infecting plants and animals, generally found in soil, water and skin. It can cause urinary tract infections, pneumonia and often attack people with weak immune systems.
Mobile phone contamination that was reported in Iran by taking random samples from different people showed a lower percentage of S. auerus (11.4%) as compared to our results that reveals the maximum presence of S. auerus. E. coli constituted 12.3% and Pseudomonas 0.3% [1].
Another study that was conducted in Bayero University, Kano; showed that 76% of the samples were contaminated with S. auerus [11]. This result agreed with our findings where S.auerus occurs in maximum number of samples (Figure 3).
Healthcare centers provides a potential risk of transmission of pathogens when the staff works in ICUs. These are harmful for the staff as well as new patients who will get exposed to different microflora. A healthcare setting study reported 53% of S. aureus, 50% of CoNS, 43% Bacillus, 18% E. coli and other isolates [12].
Most of the work done related to the mobile phone contamination focused on the infections and it’s spread in healthcare units. Our study evaluated different categories so as to cover general population as well. It represents the necessity to aware public as his device has to be managed in a way so that our cell phones don’t becaome a house for microorganisms.
Conclusion
The inability to carefully handle mobile phone had exposed the population to the risk of diseases. One should be concerned about his/her surroundings and maintain sanitation. It is very unhealthy to use the phone with dirty hands and during work. Mobile phones are source of infections that can be transmitted to other individuals. Therefore, this device should be single handed so as to avoid the cross contamination. People get exposed to new pathogens to which they are not adapted when they come in contact with the mobile phone of other individuals. Also, a periodic cleaning procedure for the cell phones should be introduced in our habit so as to eliminate the chance of spread of infections. Various disinfectants are available in the market which effectively removes the pathogens. It is always good to be hygienic and prevent the health risks. There is a need to sensitize the general population about the probable health hazards that can be caused by irresponsible handling of phone.
Acknowledgement
We thank the Director, Shriram Institute for Industrial Research, Delhi for providing the necessary facilities.
REFERENCES
- Akinyemi K, Audu A, Olabisi A, et al. The potential role of mobile phones in the spread of bacterial infections, J Infect Dev Ctries. 2009;8:628-32.
- Arora U, Devi P, Chadha A, et al. Cellphones a modern stay house for bacterial pathogens J K Sci. 2009;3:127-29.
- Brady RR, Verran J, Damani NN et al. Review of mobile communication devices as potential reservoirs of nosocomial pathogens, J Hosp Infect. 2009;71(4):295-300.
- Brady RR, Wasson A, Stirling I, et al. Is your phone bugged? The incidence of bacteria known to cause nosocomial infection in healthcare workers mobile phones, J Hosp Infect. 2006;62:123-25.
- Chauhan A, Goyal P. Isolation and Identification of Escherichia coli from various foodstuffs and their resistance against clinically significant antibiotics, J Advance in Biology. 2013;245-53.
- Chauhan A, Goyal P, Aggarwal ML, et al. Prevalence and antibiotic resistance of Bacillus strains isolated from various food stuffs. Journal of Biomedical and Pharmaceutical Research. 2013;2:8-16.
- Chauhan A, Ranjan A, Basniwal RK, et al. Probiotic, prebiotic and synbiotics in the prevention of lifestyle disorders. Int J Curr Microbiol App Sci. 2016;4:933-47.
- Chauhan A, Bharti PK, Goyal P, et al. Psychrophilic pseudomonas in antarctic freshwater lake at stornes peninsula, larsemann hills over east Antarctica. SpringerPlus 2015;4:582.
- Chauhan A, Goyal P, Varma A, et al. Microbiological evaluation of drinking water sold by roadside vendors of Delhi, India. Applied Water Science. 2017;7:1635-44.
- Chauhan A, Goyal P, Verma A, et al. In -vitro antibiotic resistance and heavy metal tolerance patterns of gram-positive and gram-negative bacteria isolated from effluent treated water of Delhi, India. Journal of Current Pharma Research. 2015;2:1449-58.
- Famurewa O, David OM. Cell phone: A medium of transmission of bacterial pathogens. World Rural Observations. 2009;1:69-72.
- Goyal P, Chauhan A, Aggarwal ML, et al. Microbiological aspects of water: Key criteria of quality. Current Research in Biological and Pharmaceutical Sciences 2012;1:57-66.