The arrow of carcinogenesis
Received: 30-Oct-2017 Accepted Date: Dec 03, 2017; Published: 15-Dec-2017
Citation: Grant SG. The arrow of carcinogenesis. J Mol Cancer. 2017;1(1):1-6.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Many models of molecular carcinogenesis involve a multi-step progression through an accumulation of genetic or epigenetic events conferring various aspects of the transformed phenotype. Identification of tissue typespecific pathways involving distinctive genes has had some success, but it is becoming clear that such pathways may well be unique to each tumor. As an alternative, some researchers have targeted the mechanisms responsible for the accumulation of aberrant genes or gene expression, the “arrows” of transition between steps. The frequency of such genetic and epigenetic changes depends on both the lifelong exposure profile for an individual, as well as host factors, such as their innate rate of replication error and ability to remediate induced DNA damage. The totality of these effects can be evaluated functionally, however, using a number of approaches. Broadly defined, cumulative determinations of somatic mutational burden have been shown to predict subsequent cancer development, as well as demonstrating the development of genomic instability as a common characteristic of aging, that presages cancer incidence. Well established and validated methodologies exist that can be applied to the monitoring of wellness in patients before cancer occurs
Keywords
DNA repair; Genomic instability; Molecular carcinogenesis; Mutational burden
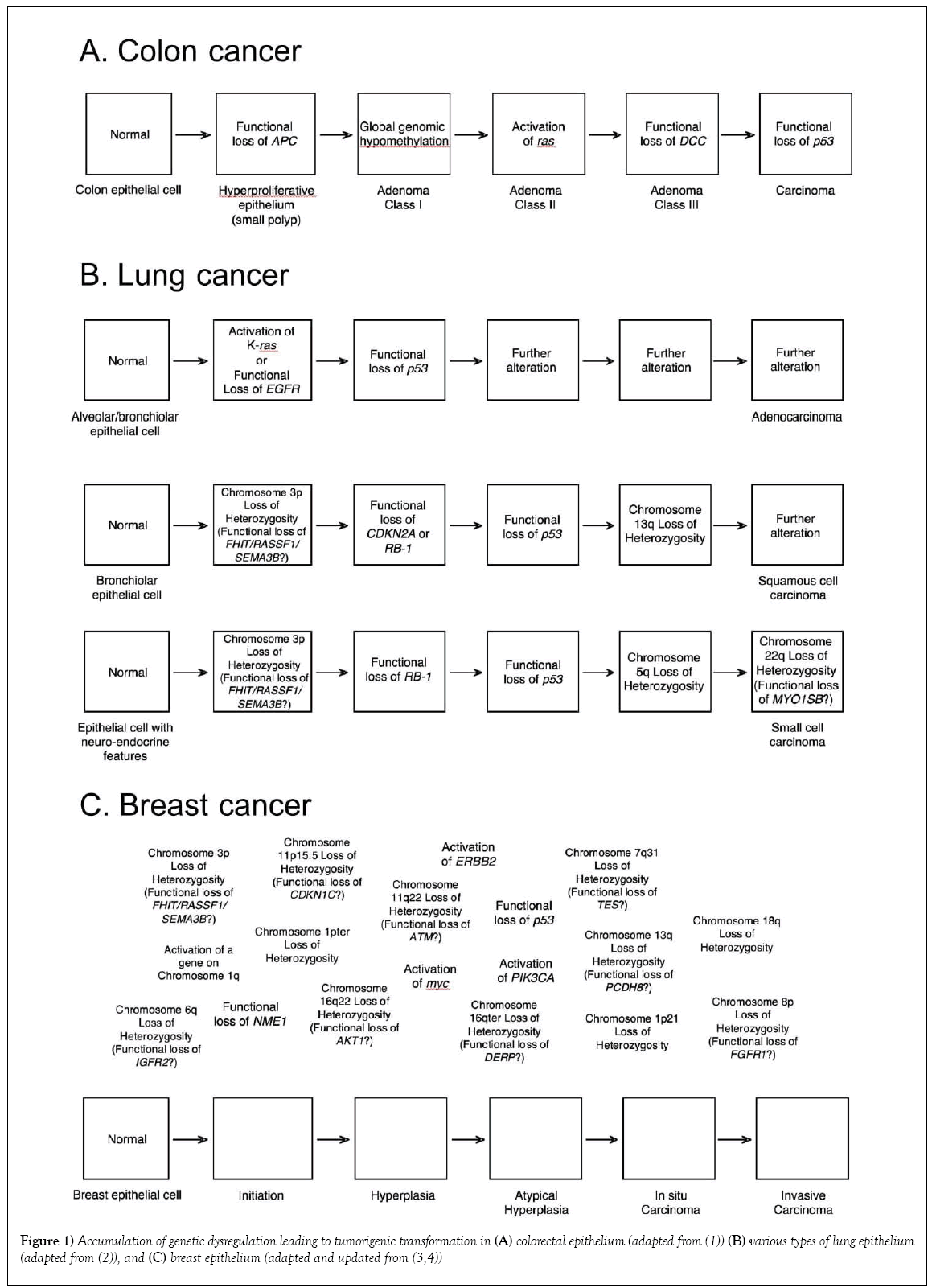
W hen first unveiled, the somatic model of colorectal cancer (Figure 1) was thought to be the harbinger of a new era of molecular cancer biology – with simple molecular pathways for each of the major cancer types that could be mined and exploited for prevention, intervention and treatment strategies. The pathway presented included both a well-established oncogene, ras, as well as the gene that exemplified tumor suppression, p53, which at that time was touted as the universal molecular constant in carcinogenesis [1-5]. Two other theoretical genes were identified by areas of consistent “loss-of-heterozygosity” at specific stages of colorectal carcinogenesis: the mutated-in-colorectal-carcinoma (MCC) gene on chromosome 5 that seemed to begin the process, and the deleted-in-colorectal-carcinoma (DCC) gene on chromosome 18, which was associated with transformation from a class II to a class III adenoma. Significant validation for the pathway was provided by the subsequent identification, cloning and characterization of both of these genes, although there is still some controversy as to whether those initially identified genes are actual the cancer “drivers” in those regions [6]. In most models (including ours), the MCC gene has been replaced with the closely linked APC gene, the genetic determinant of familial adenomatous polyposis (FAP), a hereditary syndrome with a 95% incidence of colorectal cancer by age 50 [7].
Instead of this model heralding a new era of simpler cancer molecular etiology, however, it soon became clear that colorectal cancer was a special case, with a single major molecular pathway common enough to appear to be unique. With alternate pathways (some subtype specific) and branching, most major cancer types have only nonrandom associations with specific genes, rather than established pathways. For example, there are at least three overlapping pathways for lung cancer [2] and no clear pathway or pathways have emerged for breast cancer [3,4]. Indeed, there are now three acknowledged molecular pathways for the development of colorectal cancer [8,9]. In retrospect, expecting cancer types to have unique molecular pathways was extremely naïve, since even a casual consideration of the original proposed pathway for colorectal cancer reveals functional alternatives that have since been observed. For example, dysregulation of Raf can substitute for a ras mutation, having a similar effect on the activation of the MAP kinase pathway [10,11]. Also, MDM2 acts as a sink for P53 protein, so overexpression of MDM2 is an alternative mechanism of inactivating p53 [12]. Even a single functional pathway can therefore be traversed through alternate molecular events. Thus, although the particular genes identified in Figure 1 and other proposed pathways of carcinogenesis have been used to predict risk of cancer and facilitate early detection [13,14], optimize treatment [15] and monitor treatment response [16], they require personalization for individual tumors for optimal application. There is another element to the pathway, however, that is shared by all known and hypothesized molecular models of carcinogenesis: the arrows.
ALTERNATE MECHANISMS OF HEREDITARY CARCINOGENESIS
The arrows in Figure 1 represent the mechanisms by which carcinogenesis proceeds; how the oncogenes become activated and the tumor suppressor genes inactivated. They are usually casually defined as “mutation,” although we are well aware that they include types of events that go well beyond traditional point mutation [17] (Table 1).
| Mechanisms of activation of cellular proto-oncogenes | |
|---|---|
| Mutation | Structural mutation1 to hyperactivity Regulatory mutation1 to overexpression Regulatory mutation2 to unregulated expression |
| Epigenetic activation3 | |
| Gene amplification | |
| Translocation/inversion/insertion | Functional juxtaposition of proto-oncogene coding regions into heterologous regulatory region |
| Mechanisms of inactivation of recessive tumour suppressor genes | |
| Mutation | Structural mutation2 to inactivity Regulatory mutation2 to non-expression Mutation2 affecting mRNA processing or stability |
| Epigenetic inactivation4 Gene deletion | |
| Translocation/inversion/insertion | Disruption of integrity of gene |
| Mechanisms of loss-of-heterozygosity of tumour suppressor gene | |
| Mutation | Structural mutation2 to inactivity Regulatory mutation2 to non-expression Mutation2 affecting mRNA processing or stability |
| Epigenetic inactivation4 Gene deletion | |
| Translocation/inversion/insertion | Disruption of integrity of gene |
| Chromosome loss Chromosome loss and duplication Mitotic recombination Gene conversion |
|
| 1Point mutation 2Point mutation, small deletion or insertion 3DNA hypomethylation, conformational change to euchromatin, histone deacetylation/methylation/demethylation, binding of inhibitory miRNAs 4DNA hypermethylation, conformational change to heterochromatin, histone acetylation/methylation/demethylation, binding of inductive miRNAs Revised and expanded from [17] |
|
Table 1: Molecular mechanisms of carcinogenesis
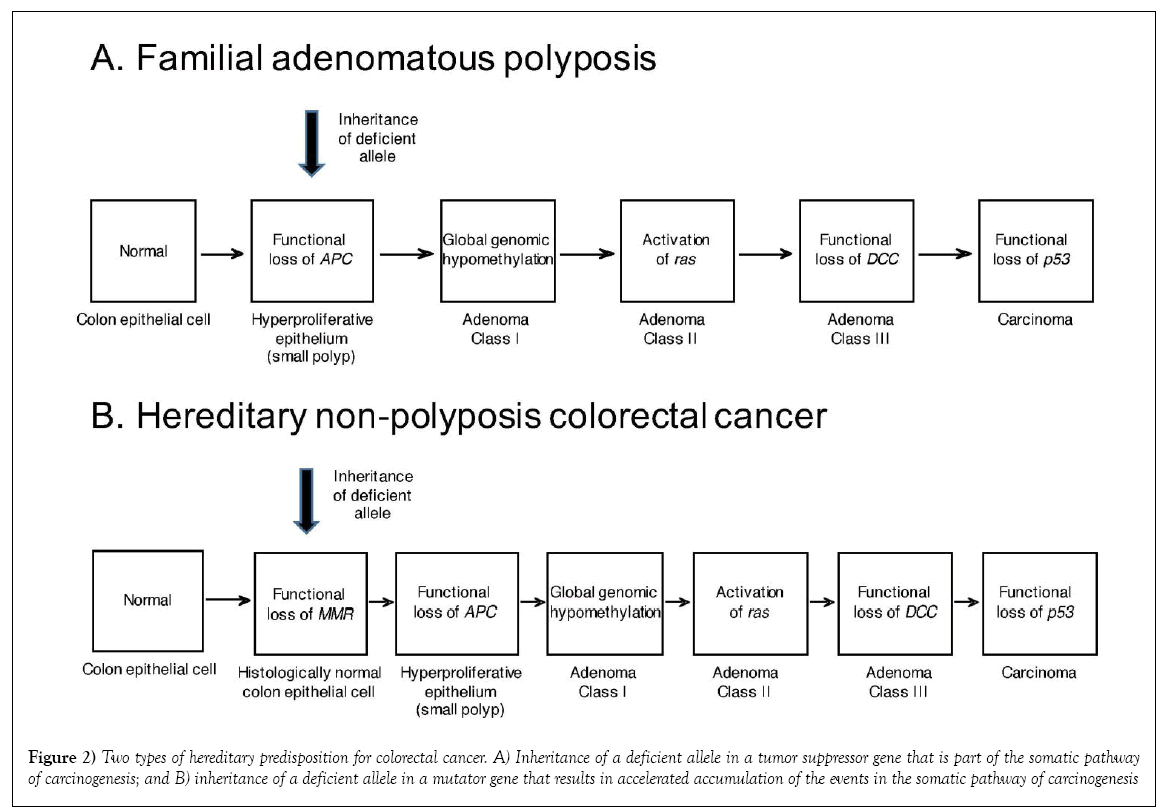
A second, and actually more common type of hereditary colorectal cancer is hereditary non-polyposis colorectal cancer (HNPCC), also known as Lynch syndrome [4,18]. Whereas, it initially appeared that the predisposition to and acceleration of onset of colorectal cancer in the other hereditary colorectal cancer syndrome, FAP, was due to inheritance of a necessary event in the common pathway of colorectal carcinogenesis, effectively shortening the pathway, HNPCC involves a set of genes not invoked in the sporadic molecular pathway, genes involved in DNA mismatch repair (MMR). The mismatch repair genes can be classified as a subset of tumour suppressor genes known as “mutator” genes; meaning that they mediate some of the mechanisms whereby accumulation of mutations can proceed to carcinogenic transformation. Indeed, although hereditary inactivating mutations in several MMR genes can contribute to carcinogenesis in specific patients and their families, in each case they still represent an additional step in the pathway of carcinogenesis (reduction to hemizygosity or homozygosity of the inherited deficient MMR gene), actually lengthening the pathway; paradoxically, however, complete loss of mismatch repair allows for a faster traverse of the pathway, with faster accumulation of the subsequent events necessary to produce the initial transformed tumor cell. Such genomic instability essentially makes the arrows themselves shorter (Figure 2).
Figure 2: Two types of hereditary predisposition for colorectal cancer. A) Inheritance of a deficient allele in a tumor suppressor gene that is part of the somatic pathway of carcinogenesis; and B) inheritance of a deficient allele in a mutator gene that results in accelerated accumulation of the events in the somatic pathway of carcinogenesis
FURTHER MECHANISMS OF GENOMIC INSTABILITY IN COLORECTAL CANCER
Another pathway of colorectal carcinogenesis has been defined largely due to its dependence on chromosomal instability (CIN) [8]. Aberrant karyotypes were one of the first characteristics of tumors to be generalized [19] and can be due to ongoing chromosomal instability [20]. Recent evidence suggests that loss of the APC gene also affects genomic stability, by inhibiting DNA base excision repair (BER) [21]. We have shown consistent downregulation of DNA nucleotide excision repair (NER) in early stage breast cancer [22]. With all of this evidence, it is difficult to understand how genomic instability was originally overlooked as a “hallmark” of cancer [23], and added later only as an “enabling factor” [24]. This is in contrast to the “mutator” theory of carcinogenesis, which states that the acquisition of events necessary for carcinogenic transformation in a single cell could not occur within a human lifetime without the increase in frequency afforded by some type of genomic instability [25]. Perhaps the resolution of these ideas would be that genomic instability is required to become cancer, but may not be required to maintain the cancer phenotype (with discussions of the possible role of genomic instability in cancer progression set aside for now).
SHARED MECHANISMS OF CARCINOGENESIS
As we have seen, there is some utility in defining the exact genes altered during carcinogenesis, even if it must be done individually for each tumour; is there any reason to similarly study the arrows? We normally think of the genetic and epigenetic mechanisms that define carcinogenesis as occurring through both endogenous processes [26] and the inevitable and unavoidable environmental, occupational, medical, lifestyle and accidental exposures we are subjected to throughout our lifetime [27]. The accumulated effect of these processes, as well as any genetic mediating factors, may be referred to as the somatic mutational “burden” and has been measured in many ways [28,29].
BLOOD-BASED EVALUATIONS OF SOMATIC MUTATIONAL BURDEN AND CANCER RISK
Most impressively, screening of baseline chromosome aberration (CA) frequency [30] or frequency of micronuclei (MN) [31] in blood cells have been shown to be predictive of subsequent cancer incidence in large prospective studies. CA analysis provided a relative risk (RR) of 1.41, with a 95% confidence interval (CI) of 1.16-1.72 [32], whereas MN had a RR of 1.53 (95%CI: 1.04-2.25) [33], both of which are statistically significant. In these studies, it is not the particular genes, but the ease of traversing a standard carcinogenic pathway, that defines cancer risk [34]. A related technique, quantitative analysis of induced chromosome breakage (also called “mutagen sensitivity”) [35], has been applied retrospectively to patients with a number of cancer types and has produced significant odds ratios that might suggest it would also be predictive for cancer (Table 2). These chromosomal analyses are available in most children’s hospitals, because they are the laboratory diagnostic standard for the cancer-prone hereditary syndrome Fanconi anemia [41,42] and a comprehensive protocol for quantitative induction of chromosome damage with a number of agents has also been recently been published [43]. Blood-based somatic mutation at the HPRT and GPA reporter genes (Table 3) has also been associated with cancer incidence in retrospective studies [44]. Odds ratios based on studies using these methods are given in Table 4. Cut-off values were determined as approximately 3 standard deviations above the mean of the control population. Note that for the GPA assay, this is similar to the criteria applied as diagnostic for the cancer-prone diseases ataxia telangiectasia [64] and Fanconi anemia [65]. These odds ratios also suggest these assays might be useful in a population screen to identify individuals at increased risk for cancer. Both of these assays have been in use experimentally for over 30 years and protocols for these procedures have also recently been published in detail [66,67]. Population studies with the GPA somatic mutation assay have shown that the incidence of high mutation frequency “outliers,” perhaps indicative of the development of genomic instability in bone marrow stem cells, rises exponentially beginning at age 45 [68]. Notably, this parallels the age of incidence of many solid tumors [69]. Perhaps acquisition of genomic instability is the defining step in carcinogenesis, when development of a fully transformed cell becomes inevitable? Population screening and monitoring via application of one or more of these assays of the arrow of carcinogenesis should become a regular part of the evaluation of disease risk in asymptomatic human beings, rather than wait for the development of overt cancer.
| Cancer type | Inducing agent | Odds Ratio (95% Confidence Interval) |
References |
|---|---|---|---|
| Lung | Benzo[a]pyrene diol epoxide Bleomycin |
2.15 (1.39-3.33) 2.69 (1.44-5.04) |
[36,37] |
| Head and neck | Benzo[a]pyrene diol epoxide | 1.56 (1.27-1.91) | [38] |
| Breast | Bleomycin | 2.8 (1.7-4.5) | [39] |
| Melanoma | 4-nitroquinoline-1-oxide | 1.78 (1.12-2.84) | [40] |
Table 2: Association of elevated mutagen sensitivity (induced chromosome aberrations) with cancer
| Features | Events detected | |
|---|---|---|
| HPRT | Well-established assay, with extensive normal database Applicable to everyone except patients with Lesch-Nyhan syndrome (hereditary deficiency of HPRT) ~20 mL of fresh blood required Expensive and labor-intensive: requires cell culture and clonogenic drug selection Mutant colonies can be genetically analyzed: generate “mutational spectra” Comparable in vitro, animal versions |
Gene-specific mutation1 Structural mutation to inactivity Regulatory mutation to non-expression Mutation affecting mRNA processing or stability Epigenetic inactivation Gene deletion Translocation/inversion/insertion Disruption of integrity of gene |
| GPA | Well-established assay, with extensive normal database <1 mL of fresh blood required Inexpensive and rapid: direct flow cytometric detection of mutants Mutant phenotype cannot be conformed at the DNA level Resolution of mutants with allele-loss and loss-and-duplication phenotypes Loss-and-duplication phenotype can be resolved only in GPA (MN) heterozygotes (~50% population) |
MutationStructural mutation to inactivityRegulatory mutation to non-expressionMutation affecting mRNA processing or stability Epigenetic inactivation Gene/chromosome deletion Translocation/inversion/insertionDisruption of integrity of gene, position effects Chromosome loss Chromosome loss and duplication Mitotic recombination Gene conversion |
| 1Due to location on X chromosome, limited ability to detect extragenic mechanisms Updated and expanded from (34) |
||
Table 3: The HPRT and GPA in vivo somatic mutation assays
| HPRT somatic mutation assay | ||||
|---|---|---|---|---|
| Population | N | N Mf ≤ 2 × 10-5 |
N Mf > 2 × 10-5 |
Odds Ratio (95% CI) |
| All cancer patients1 | 92 | 84 | 8 | 5.12 (2.03-12.88) |
| Disease-free controls2 | 657 | 645 | 12 | |
| GPA somatic mutation assay | ||||
| Population | N | N Mf ≤ 3 × 10-5 |
N Mf > 3 × 10-5 |
Odds Ratio (95% CI) |
| All cancer patients3 | 98 | 63 | 35 | 4.24 (2.66-6.75) |
| Disease-free controls4 | 802 | 709 | 93 | |
| 1Data from cervical, bladder, endometrial, lung, head and neck, colon and prostate cancer [45], Hodgkin’s disease, Ewing sarcoma and rhabdomyosarcoma [46], breast cancer [47] and lymphoma and breast, esophageal, stomach, colorectal, lung, ovarian, testicular, choriocarcinoma, squamous cell carcinoma and bladder cancer [48] 2Data from controls from cancer studies [46-48] and population studies [49-56] 3Data from adenocarcinoma, bone metastases, brain, breast, colon, germ cell, head and neck, Hodgkin’s and non-Hodgkin’s lymphoma, melanoma, renal, neuroblastoma, ovarian, liver, lung, cervical and thoracic cancers [57,58], testicular cancer [59] and breast cancer [60] 4Data from controls from cancer studies [57,60] and population studies [61-63] N: Number; Mf: Mutation frequency |
||||
Table 4: Calculation of odds ratios for cancer retrospective studies of mutation frequency based on reporter genes
REFERENCES
- Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319: 525-32.
- Yatabe Y, Borczuk AC, Powell CA. Do lung adenocarcinomas follow a stepwise progression? Lung Cancer. 2011;74:7-11.
- Bièche I, Lidereau R. Genome-based and transcriptome-based molecular classification of breast cancer. Curr Opin Oncol. 2011; 23:93-9.
- Guedj M, Marisa L, de Reynies A, et al. A refined molecular taxonomy of breast cancer. Oncogenomics. 2012;31:1196-1206.
- Finlay CA. p53 loss of function: implications for the processes of immortalization and tumorigenesis. BioEssays. 1992;14:557-60.
- Ji X, Tang J, Halberg R, et al. Distinguishing between cancer driver and passenger gene alteration candidates via cross-species comparison: a pilot study. MBC Cancer. 2010;10:426.
- Kanth P, Grimmett J, Champine M, et al. Hereditary colorectal polyposis and cancer syndromes: A primer on diagnosis and management. Am J Gastroenterol 2017;112:1509-25.
- Mundade R, Imperiale TF, Prabhu L, et al. Genetic pathways, prevention and treatment of sporadic colorectal cancer. Oncoscience. 2014;1:400-6.
- Bae JM, Kim JH, Kang GHK. Molecular subtypes of colorectal cancer and their clinicopathologic features, with an emphasis on the serrated neoplasia pathway. Arch Pathol Lab Med. 2016;140:406-12.
- Fransén K, Klintenäs M, Österström A, et al. Mutation analysis of the BRAF, ARAF and RAF-1 genes in human colcorectal adenocarcinomas. Carcinogen. 2004;25:527-33.
- Nie F, Cao J, Tong J, et al. Role of Raf-kinase inhibition protein in colorectal cancer and its regulation by hydroxycamptothecine. J Biomed Sci. 2015;22:56.
- Hata AN, Rowley S, Archibald HL, et al. Synergistic activity and heterogeneous acquired resistance of combined MDM2 and MEK inhibition in KRAS mutant cancers. Oncogene. 2017;36:6581-91.
- Mak T, Lalloo F, Evans DGR, et al. Molecular stool screening for colorectal cancer. Br J Surg. 2004;91:790-800.
- Lim SH, Becker TM, Chua W, et al. Circulating tumour cells and circulating free nucleic acid as prognostic and predictive biomarkers in colorectal cancer. Cancer Lett. 2014;346:24-33.
- Punt CJA, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14:235-46.
- Barault L, Amatu A, Siravegna G, et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut. 2017.
- Grant SG, Green DM, D’Angio GJ. Mutation, segregation and childhood cancer. In Late Effects of Treatment for Childhood Cancer. New York: Wiley-Liss. 1992:121-32.
- Da Silva FC, Wernhoff P, Dominguez-Barrera C, et al. Update on hereditary colorectal cancer. Anticancer Res. 2016;36:4399-4406.
- Wolman SR. Karyotypic progression in human tumors. Cancer Metastasis Rev. 1983;2:257-93.
- Jefford CE, Irminger-Finger I. Mechanisms of chromosome instability in cancers. Crit Rev Oncol Hematol. 2006;59:1-14.
- Narayan S, Sharma R. Molecular mechanism of adenomatous polyposis coli-induced blockade of base excision repair pathway in colorectal carcinogenesis. Life Sci. 2015;139:145-52.
- Latimer JJ, Johnson JM, Kelly CM, et al. Nucleotide excision repair deficiency is intrinsic in sporadic stage I breast cancer. Proc Natl Acad Sci U S A. 2010;107:21725-30.
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;144:57-70.
- Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646-74.
- Fox EJ, Prindle MJ, Loeb LA. Do mutator mutations fuel tumorigenesis? Cancer Metastasis Rev. 2013;32:353-61.
- Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001; 477:7-21.
- Wogan GN, Hecht SS, Felton S, et al. Environmental and chemical carcinogenesis. Semin Cancer Biol. 2004;14:473-86.
- Grant SG, Jensen RH. Use of hematopoietic cells and markers for the detection and quantitation of human in vivo somatic mutation. In: Garratty G, ed. Immunobiology of Transfusion Medicine. Marcel Dekker, New York. 1993;299-323.
- Møller P. Genotoxicity of environmental agents assessed by the alkaline comet assay. Basic Clin Pharmacol Toxicol. 2005;96:1-42.
- Hagmar L, Brøgger A, Hansteen IL, et al. Cancer risk in humans predicted by increased levels of chromosomal aberrations in lymphocytes: Nordic study group on the health risk of chromosome damage. Cancer Res. 1994;54:2919-22.
- Murgia E, Ballardin M, Bonassi S, et al. Validation of micronuclei frequency in peripheral blood lymphocytes as early cancer risk biomarker in a nested case-control study. Mutat Res. 2008;639:27-34.
- Bonassi S, Norppa H, Ceppi M, et al. Chromosome aberration frequency in lymphocytes predicts the risk of cancer: Results from a pooled cohort study of 22,358 subjects in 11 countries. Carcinogen. 2008;29:1178-83.
- Bonassi S, Znaor A, Ceppi M, et al. An increased micronucleus frequency in peripheral blood lymphocytes predicts the risk of cancer in humans. Carcinogen. 2007;28:625-31.
- Grant SG. Translating mutagenesis into carcinogenesis. J Carcinogen Mutagen. 2012;3:e106.
- Wu X, Gu J, Spitz MR. Mutagen sensitivity: A genetic predisposition factor for cancer. Cancer Res. 2007;67:3493-5.
- Li D, Firozi PF, Wang LE, et al. Sensitivity to DNA damage induced by benzo(a)pyrene diol epoxide and risk of lung cancer: A case-control analysis. Cancer Res. 2001;61:1445-50.
- Zheng YL, Loffredo CA, Yu Z, et al. Bleomycin-induced chromosome breaks as a risk marker for lung cancer: A case-control study with population and hospital controls. Carcinogenesis. 2003;24:269-74.
- Wang LE, Xiong P, Zhao H, et al. Chromosome instability and risk of squamous cell carcinomas of head and neck. Cancer Res. 2008; 68:4479-85.
- Kosti O, Byrne C, Meeker KL, et al. Mutagen sensitivity, tobacco smoking and breast cancer risk: A case-control study. Carcinogenesis. 2010;31:654-59.
- Wang LE, Li C, Xiong P, et al. 4-nitroquinoline-1-oxide-induced mutagen sensitivity and risk of cutaneous melanoma: A case-control analysis. Melanoma Res. 2016;26:181-87.
- Auerbach AD. Diagnosis of Fanconi anemia by diepoxybutane analysis. Curr Protoc Hum Genet. 2015;85:1-17
- Francies FZ, Wainwright R, Poole J, et al. Diagnosis of Fanconi anemia by ionizing radiation- or mitomycon C-induced micronuclei. DNA Repair. 2017;61:17-24.
- Wu X, Zheng YL, Tsu TC. Mutagen sensitivity as measured by induced chromatid breakage as a marker of cancer risk. Methods Mol Biol 2014;1105:183-92.
- Grant SG. Molecular epidemiology of human cancer: biomarkers of genotoxic exposure and susceptibility. J Environ Pathol Toxicol Oncol. 2001;20:245-61.
- Ammenheuser MM, Au WW, Whorton EB Jr, et al. Comparison of Hprt variant frequencies and chromosome aberration frequencies in lymphocytes from radiotherapy and chemotherapy patients: A prospective study. Environ Mol Mutagen. 1991;18:126-35.
- Lange BJ, Prantner JE. The emergence of 6-thioguanine-resistant lymphocytes in pediatric cancer patients. Mutat Res. 1982;94:487-99.
- Messing K, Bradley WEC. In vivo mutant frequency rises among breast cancer patients after exposure to high doses of g-radiation. Mutat Res. 1985;152:107-12.
- Dempsey JL, Seshadri RS, Morley AA. Increased mutation frequency following treatment with cancer chemotherapy. Cancer Res. 1985;45:2873-7.
- Tates AD, van Dam FJ, van Mossel H, et al. Use of the clonal assay for the measurement of frequencies of Hprt mutants in T-lymphocytes from five control populations. Mut Res. 1991;253:199-213.
- Branda RF, Sullivan LM, O’Neill JP, et al. Measurement of Hprt mutant frequencies in T-lymphocytes from healthy human populations. Mutat Res. 1993;285:267-79.
- Jones IM, Moore DH, Thomas CB, et al. Factors affecting Hprt mutant frequency in T-lymphocytes of smokers and nonsmokers. Cancer Epidemiol Biomarkers Prev. 1993;2:249-60.
- Finette BA, Sullivan LM, O’Neill JP, et al. Determination of Hprt mutant frequencies in T-lymphocytes from a healthy pediatric population: Statistical comparison between new-born, children and adult mutant frequencies, cloning efficiency and age. Mutat Res. 1994;308:223-31.
- Hüttner E, Holzapfel B, Kropf S. Frequency of Hprt mutant lymphocytes in a human control population as determined by the T-cell cloning procedure. Mutat Res. 1995;348:83-91.
- Becker R, Nikolova T, Wolff I, et al. Frequency of Hprt mutants in humans exposed to vinyl chloride via an environmental accident. Mutat Res. 2001;494:87-96.
- Finnette BA, Kendall H, Vacek PM. Mutational spectral analysis at the HPRT locus in healthy children. Mutat Res. 2002;505:27-41.
- Evdokimova VN, Babra B, Grant SG. Cell viability as evaluated by cloning efficiency is the major factor affecting in vivo Hprt somatic mutation frequencies. Environ Mol Mutagen. 2004;44:198.
- Bigbee WL, Wyrobek AW, Langlois RG, et al. The effect of chemotherapy on the in vivo frequency of glycophorin A ‘null’ variant erythrocytes. Mutat Res 1990; 240:165-75.
- Grant SG, Bigbee WL. Bone marrow somatic mutation after genotoxic cancer therapy. Lancet. 1994;343:1507-8. Erratum: Lancet. 1994;344:415.
- Perera FP, Motzer RJ, Tang D, et al. Multiple biological markers in germ cell tumor patients treated with platinum-based chemotherapy. Cancer Res. 1992;52:3558-65.
- Grant SG, Myers NT, Kelley JL III, et al. Longitudinal somatic mutational biomonitoring of genotoxic breast cancer chemotherapy reveals considerable inter-individual variability in bone marrow response with potential clinical significance. Proc Am Assoc Cancer Res. 2006;47:461.
- Jensen RH, Bigbee WL, Langlois RG, et al. Laser-based flow cytometric analysis of genotoxicity of humans exposed to ionizing radiation during the Chernobyl accident. SPIE Proc. 1990; 1403, 372-80.
- Grant SG, Bigbee WL, Langlois RG, et al. Methods for the detection of mutational and segregational events: relevance to the monitoring of survivors of childhood cancer. In: Late Effects of Treatment for Childhood Cancer, (Green DM, D’Angio GJ, eds.), Wiley-Liss, New York. 1992;133-50.
- Grant SG, Bigbee WL. Genetic and environmental factors affecting lifetime human somatic mutation and segregation frequencies beginning in utero. Am J Med Genet. 1994;52:367.
- Grant SG, Reeger W, Wenger SL. Diagnosis of ataxia telangiectasia with the glycophorin A somatic mutation assay. Genet Testing. 1998;1:261-7.
- Evdokimova VE, McLoughlin RK, Wenger SL, Grant SG. Use of the glycophorin: A bone marrow somatic mutation assay for rapid, unambiguous identification of Fanconi anemia homozygotes regardless of GPA genotype. Am J Med Genet. 2005;135A:59-65.
- Hou SM. The human T-cell cloning assay: Identifying genotypes susceptible to drug toxicity and somatic mutation. Methods Mol Biol. 2014;1105:283-301.
- Myers NT, Grant SG. The blood-based glycophorin A human in vivo somatic mutation assay. Methods Mol Biol. 2014;1105:193-202.
- Bigbee WL, Fuscoe JC, Grant SG, et al. Human in vivo somatic mutation measured at two loci: individuals with stably elevated background erythrocyte glycophorin A (gpa) variant frequencies exhibit normal T-lymphocyte hprt mutant frequencies. Mutat Res. 1998;397:119-36.
- Global Burdon of Disease Cancer Collaboration. Global, regional and national cancer incidence, mortality, years of life lost, years lived with disability and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3:524-48.