The popular 3675SGF deletion in nsp6 domain of ORF1ab polyprotein of SARS-CoV-2 was spread into Alpha, Beta, Gamma, Iota and Omicron variants except highly infectious and death promoting Delta variant
Received: 16-Feb-2023, Manuscript No. puljmbr-23- 6248; Editor assigned: 18-Feb-2023, Pre QC No. puljmbr-23- 6248 (PQ); Accepted Date: Mar 07, 2023; Reviewed: 01-Mar-2023 QC No. puljmbr-23- 6248 (Q); Revised: 05-Mar-2023, Manuscript No. puljmbr-23- 6248 (R); Published: 10-Mar-2023, DOI: 10.37532/puljmbr.2023.6(2).41-44
Citation: Chakraborty AK. The popular 3675SGF deletion in Nsp6 domain of ORF1ab polyprotein of SARS-CoV-2 was spread into alpha, beta, gamma, iota and omicron variants except highly infectious and death promoting delta variant. J Mic Bio Rep. 2023;6(2):41-44.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
The SARS-CoV-2 was appeared in mid-2020 but the first RNA sequence was available since December, 2020. The higher transmission and disease severity were seen since acquisition of D614G dominant point mutation in spike protein as well as P4715L point mutation in the RNA-dependent RNA polymerase (RdRp). The next event occurred in B.1.1.7 Alpha variant was acquisition of N501Y point mutation and 69HV two AAs deletion in the spike protein. Interestingly, 3675SGF three AAs deletions were also occurred in ORF1ab polyprotein in B.1.1.7 variants. The appearance of notorious Delta variant in Mid-2021 was shocking as too many deaths happened worldwide. The Delta variants (B.1.617.2 and AY.X) acquired 157FR two AAs deletion instead 69HV in B.1.1.7 variant. Here, we showed that Delta variant was spread worldwide but has no 3675SGF deletion. Moreover, 3675SGF deletion was found in all Omicron variants which were appeared in December, 2021 and its subvariants BA.2.75, BA.5.2.1.7 (BF.7), BQ.1, BQ.1.1 and XBB.1.5 which were spreading now worldwide with mild infections. Thus, Delta variant has very complete ORF1ab protein (9096 AAs) to attend high titre and to infect more human lung cells. Further, 119DF deletion in ORF8 activator protein and 157FR deletion in spike might be contributed to higher pathogenicity in Delta including P2046L and P2287S mutations in nsp3 main protease, P4715L and G5063S mutations in RdRp and P6128S and A6319V mutations in nsp14 Ribonuclease. Further, complete attenuation of Delta corona virus spread recently likely due to herd immunity and wide spread inoculation with COVID-19 vaccine.
Key Words
SARS-CoV-2, Delta variant, SGF deletion, Nsp6, ORF1ab polyprotein, Higher transmission, High titre COVID-19
Introduction
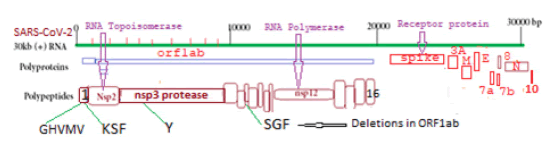
The COVID-19 virus is a ~30kb single-stranded positive-sense large RNA virus which infects human lung cells to cause corona disease since 2020 [1]. COVID-19 is related to six different coronaviruses like CoV229E, CoV-HKU1, CoV-OC43, CoV-NL63, SARS-CoV, and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) which were known since 2003-2012 [2,3]. Within two years the coronavirus acquired many mutations and deletions with the difference in its pathogenicity and severity to cause death [4-8]. Five VOCs of SARS-CoV-2 mainly caused millions of death worldwide and named B.1.1.7 (Alpha; U.K.), B.1.351 (Beta; South Africa), P.1 (Gamma; Brazil), B.1.617.2 (Delta; India), and B.1.1.529 (Omicron; USA, Africa). Analysis suggested that spike protein (1273AA) of COVID-19 had gone through extensive mutations and deletions than large polyprotein ORF1ab (7096aa) which was degraded into sixteen polypeptides like RNA topoisomerase (nsp2), proteases (nsp3 and nsp5), RNA-dependent RNA polymerase (nsp12), RNA helicase-capping methyltransferase (nsp13), RNases (nsp14 and nsp15) and 2’-uridine methyltransferase (nsp16) [9-13]. The different peaks of VOCs appeared at different times and stayed for some time but a new wave appeared due to the antigenic shift of COVID-19 usually due to spike protein mutations [14,15]. Here, we showed that more severe form of COVID-19, Delta corona
Materials and Methods
We searched PubMed to get an idea of published papers on ORF1ab and also searched the SARS-CoV-2 NCBI database using BLAST-N and BLAST-X search methods to get sequences. Multi-alignment of protein was done by MultAlin software and multi-alignment of DNA by CLUSTAL-Omega software, EMBL-EBI [15-20]. The ORF1ab mutants were obtained by BlastN search of deletion boundary of 60-100nt sequence and then analyzing the sequences with 95%-100% similarities [21]. The other ORF1ab mutants were detected by Blast-N search and Blast-X- search with selected deletion boundaries. The hairpin structure of ~ 120-200nt sequence was done by Oligo Analyzer 3.1 software (Integrated DNA Technologies). The protein 3-D structure was determined by SWISS-Model software with normal vs. mutant peptides [22,23].
Results
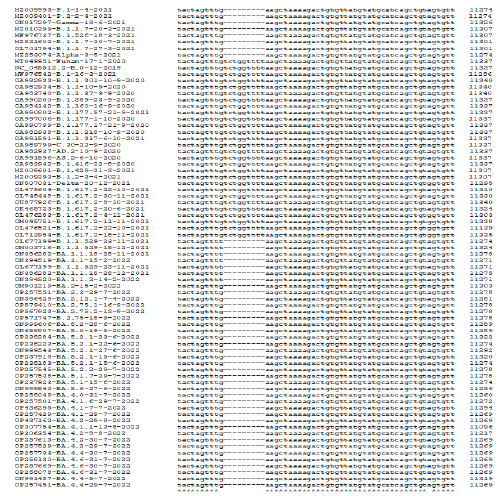
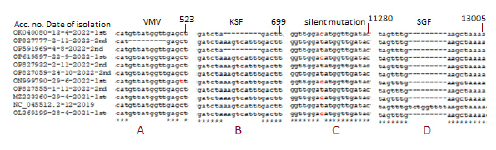
Multi-alignment was a powerful tool to compare DNA, RNA, and protein sequences from different sources to see mutations and deletions of RNA viruses like SARS-CoV-2. The multi-alignment of 30kb genomes gave 100 pages of data and thus we only showed the representative area with deletions and point mutations (Figure 1). There were hundreds of silent mutations genome-wide but unless there was an amino acid change, title effects might occur. Figure 2 showed a multi-alignment portion of different COVID-19 sequences where SGF deletion (5’-TCTGGTTTT-3’) occurred. We had given accession numbers, type of variant, and date of coronavirus isolation from patients starting Wuhan virus (B.0 variant) which was available first in December 2020. The SGF deletion was not prominent in early lineages like B.1 and B.1.2, as well as higher variants like B.1.160, B.1.389, B.1.177, B.1.177.17, B.1.177.218, B.1.416, B.1.429 including B.1.1.37, B.1.1.301 and B.1.1.317 but surprisingly B.1.1.7 variant and B.1.526 variant, had SGF deletion. More surprising fact there was no SGF deletion in B.1.617.2 Delta variant (accession numbers: OL475604, OL745448, OU877926, OK465723, OM045751, OQ121361, OQ119975, OQ119976, OQ121585, OQ121550, OQ099094 and OQ099096) (Figure 2). The total amino acids of ORF1ab polyprotein varied in different important COVID-19 variants and subvariants. For example, Wuhan=9096 AAs as also Delta variant. But three amino acid deletions (3675SGF) were found in ORF1ab protein (nsp6 protein region) of Alpha and Omicron BA.2/BA.5 (ORF1ab=7093 AAs) but at the same region 3674LSG deletion, as well as extra 2083S deletion, were found in Omicron BA.1 coronavirus (ORF1ab=7092 AAs). Whereas, extra three amino acids (141KSF) deletions were found in Omicron BA.4 variant (ORF1ab=7090 AAs) and such change was utilized to detect BA.4 Omicron variant by BLAST-2 alignment with oligonucleotide selected at the deletion boundary [24]. We multi-aligned 40 sequences of Omicron BA.1, BA.2, BA.4, and BA.5 subvariants including B.1.1.529 to demonstrate SGF deletion happened in all these Omicron variants including recently appeared B.2.75, BF.7, BQ.1 and XBB.1 (Figure 2). Truly, before the appearance of Omicron variants, B.1.617.2 and AY.X were the major coronavirus population but no SGF deletion was detected! On the other hand, all Omicron populations carried such SGF deletion! Such special distribution of SGF deletion suggested some way Omicron was derived from B.1.1.7 (Alpha) population or related B.1.526 (Iota) and P.1 (Gamma) but not B.1.351 (Beta). We thought B.1.1.7 may the source of the Omicron BA.1 variant because 69HV deletion in the spike appeared in B.1.1.7 first but Omicron BA.2 had no such 69HV deletion and that way such a variant may be created from P.1 or B.1.526 variant. However, the creation of Omicron~20 mutations in the RBD domain of spike protein likely happened through a thousand recombination and point mutations
Figure 2: Multi-alignment to show the 3675SGF deletion was found in Alpha, Beta, Gamma, and Iota variants as well as in all Omicron subvariants but not in Delta variants. (Alpha=B.1.1.7; Beta=B.1.351; Gamma=P.1; Iota=B.1.526; Delta=B.1.617.2 and AY.103; Omicron=BA.1, BA.2, BA.4, BA.5, BF.7, XBB.1.5, BQ.1, BN.1 etc).
Figure 3 demonstrated the composite data of multi-alignment to demonstrate other minor deletions and point mutations in the ORF1ab like at the 82GHVMV locus (accession no. OP827777) and 141KSF locus (accession no. OP591969) in the nsp1 protein and other sites like silent T>C mutation at 11269 positions. Now the question arises then what may be other factors for the severe pathogenicity of the Delta variant? We BLAST-2 aligned spike protein of Wuhan and Delta to see the important mutations as shown in Figure 4. We found two important mutations L452R (450R in Delta) and T478K (476K in Delta) in the RBD domain of the spike. At least the L452 mutation was found involved in immune escape and higher transmission [25, 26]. However, major spike mutations (T19R, T95I, G142D, E156G) and deletions (157FR) were located in the NH2 terminus (1-180 AAs) (Figure 4).
Figure 3: Detection of ORF1ab other deletions in SGF deletion SARS-CoV-2 mutants as well as T>C silent mutation (GAT=GAC=D; no AA change) at 11269 positions
Figure 4: The Spike protein mutations (nine AAs point mutations and two AAs deletions) in the delta variant. The sequences were derived from accession numbers NC_045512.2 (Wuhan; protein id: YP_009724390) and OM542166 (Delta; protein id: UKM99895). Parts of the alignment were shown. Major changes were found in 1-180 AAs and in the RBD domain L452R and T478K two important mutations needed for higher pathogenicity. The important 157FR two AAs deletions were shown with adjacent E156G point mutation. The D614G dominant mutation was also shown that involved in higher transmission in presence of important R4715L RdRp mutation in ORF1ab polyprotein (not shown here).
We also demonstrated the 119DF deletion in the Delta variant (Figure 5). Such deletion in Delta sequences found in the database suggested truncated non-functional ORF8 trans-activator protein which interacted with many accessory cellular proteins like histones, MHC-1, and interferons [27-29]. Interestingly, ORF8 protein was also truncated in B.1.1.7 variant which also caused severe corona diseases (data not shown). In Figure 6, we showed the two important mutations (P4715L and G5063S) in the RNAdependent RNA Polymerase enzyme required for higher transmission and disease severity. Similarly, BLAST-2 similarity between Wuhan and Delta ORF1ab proteins showed two important mutations (P2046L and P2287S) in the nsp3 main protease. We know alterations of two Proline might be responsible for efficient protease activities needed for better viral assembly. We also showed the two important mutations (P6128S and A6319V) in the nsp14 ribonuclease of the Delta variant where the specificity of nuclease to host specific mRNAs might be changed. In truth, virus-host interactions select the disease severity, viral immunity, and virus clearance (Figures 7a and 7b).
Figure 5: The delta variant had important 119DF two AAs deletion at the end of the ORF8 protein which might be involved in an increase in pathogenicity. Note that another VOC B.1.1.7 COVID-19 also had truncated ORF8 protein in its genome but other VOCs like Beta, Gamma, and Epsilon, etc had normal length ORF8 protein.
Figure 6: BLAST-2 alignment of 266-21555 Wuhan sequence (NC_045512.2) with Delta sequence (accession no. OM542166). Two mutations (P4715L and G5063S) were shown for RNA-dependent RNA Polymerase involved in higher transmission and disease severity. Total of 20 mutations were found in the ORF1ab gene but few were silent as in RNA topoisomerase T>C mutation (AAT=AAC) at 2049 coding Asparagine.
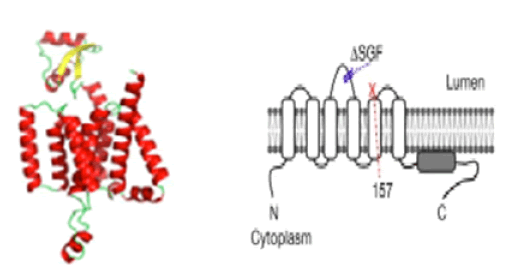
We attempted to analyze the 3-D model structure of the SGF mutant of nsp6 protein but we found no crystal structure of normal nsp6 was solved yet. However, running SWISS-Model we found a mere 20% homology at the C-terminus with a hypothetical protein TT1805 from Thermus thermophilus HB8 and E. coli or S. aureus ribosomal protein S16 (ID: 6q97.1.P and 5t7v.1.F respectively) and model was incomplete (data not shown). Pandey et al gave some model structures of nsp6 protein with seven transmembrane domains using AlphaFold software (Figure 8) [30]. The study indicated in tissue culture cells that SGF-deleted nsp6 may form better phagosomes in the replication complex with or without nsp3 and nsp4 proteins [31]. Thus, more work is necessary to solve the function of SGF deletion mutants. Nevertheless, we speculated the other mutations in RdRp, protease, and ribonuclease of the Delta variant to attend high titer, higher transmission, and possibly high risk of life.
Discussion
The Alpha, Beta, Gamma, and Delta coronaviruses were spread maximum at certain time points but were diminished due to viral herd immunity as well as rapid worldwide inoculation with spike vaccine which had shown to be best protective with high antibody titer [32]. We demonstrated a few genetic changes that surely gave high titer and severe disease patterns by Delta coronaviruses (Figure 2 and Figure 3). Such viruses form syncytia with human lung cells through ACE-2 receptors destroying cells [33]. However, the Omicron virus spread was maximum now and earlier human sera infected with Delta, Alpha, Beta, and Gamma corona viruses failed to cure Omicron virus-infected patients [34]. Surely, a clear demonstration of the absence of SGF deletion in the nsp6 protein of Delta coronaviruses was an important observation. The nsp6 protein forms smaller phagosomes at the host membrane of lung cells for virus assembly and internalization and such an autophagy process is important for the virus life cycle [35]. Similarly, ORF8 protein 119DF deletion mutant in Delta coronavirus was another important fact (Figure 5). Many small accessory proteins like nsp6, nsp9, ORF3, ORF7a, and ORF8 were involved in important functions in COVID-19 spread and viral immunity, and more and more work and Database analysis needed for coronavirus control [36-38]. Interestingly, more and more spliced mRNAs were detected in COVID-19-infected cells and more small trans-activator proteins may be discovered soon demonstrating the clear mechanism of coronavirus disease [39]. The complete structure of ORF1ab polyprotein contributed to higher transmission with higher titer in the Delta variant causing high fatality. If terminal 2 AAs deletion (219DF) in the ORF8 protein of Delta variant has any effect on pathogenicity is not clear yet. The deletions of amino acids Asp119 and Phe120 in ORF8 of the delta variant resulted in structural instability of ORF8 dimer (PDB ID:7JTL) caused by disruption of hydrogen bonds and salt bridges as revealed by structural analysis and MD simulation studies [40].
Conclusion
MHC-1 interactions may be hindered causing a better immune response by the host.
References
- Wu F, Zhao S, Yu B, et al. Complete genome characterisation of a novel coronavirus associated with severe human respiratory disease in Wuhan, China. BioRxiv. 2020.
- Lu G, Wang Q, Gao GF. Bat-to-human: spike features determining ‘host jump’of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends in microbiology. 2015;23(8):468-78.
- Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. New England journal of medicine. 2003;348(20):1953-66.
- Chakraborty AK. The 249RWMD spike protein insertion in Omicron BQ. 1 subvariant compensates the 24LPP and 69HV deletions and may cause severe disease than BF. 7 and XBB. 1 subvariants.2023.
- Ge XY, Li JL, Yang XL, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535-8.
- Liu Z, Zheng H, Yuan R, et al. Identification of a common deletion in the spike protein of SARS-CoV-2. BioRxiv. 2020:2020-03.
- Meng B, Kemp SA, Papa G, et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha Variant B. 1.1. 7. Cell reports. 2021;35(13):109292.
- Chakraborty AK, Chanda A. New Biotechnological Exploration on COVID-19 Proteins: Functions, Mutational Profiles and Molecular Targets for Drug Design. Sun Text Rev Virol. 2021;2(1):115.
- Chakraborty AK. Coronavirus Nsp2 protein homologies to the bacterial DNA topoisomerase I and IV suggest Nsp2 protein is a unique RNA topoisomerase with novel target for drug and vaccine development.
- Noske GD, Nakamura AM, Gawriljuk VO, et al. A crystallographic snapshot of SARS-CoV-2 main protease maturation process. Journal of Molecular Biology. 2021;433(18):167118.
- Chakraborty AK. Clinical, Diagnostic and Therapeutic Implications of Coronavirus ORFab Polyprotein Associated Nsp16 Protein-A Bioinformatics Approach. Acta Scientific Medical Sciences. 2020;4(5):97-103.
- Gobeil SM, Janowska K, McDowell S, et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science. 2021;373(6555): eabi6226.
- Chakraborty AK. Multi-Alignment Compariso n of Coronavirus Non-Structural Proteins Nsp13-Nsp1 6 with Ribosomal Proteins and other DNA/RNA Modifyin g Enzymes Suggested their Roles in the Regulation of Host Protein Synthesis. Int J Clin Med Info. 2020;3(1):7-19.
- McCarthy KR, Rennick LJ, Nambulli S, et al. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371(6534):1139-42.
- Sixto-Lopez Y, Correa-Basurto J, Bello M, et al. Structural insights into SARS-CoV-2 spike protein and its natural mutants found in Mexican population. Scientific Reports. 2021;11(1):4659.
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool J Mol Biol. 1990;215(3):403-10.
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic acids research. 1988;16(22):10881-90.
- Yang Y, Jiang XT, Zhang T. Evaluation of a hybrid approach using UBLAST and BLASTX for metagenomic sequences annotation of specific functional genes. PLoS One. 2014;9(10): e110947.
- Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of highâ?quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7(1):539.
- Wallace IM, Blackshields G, Higgins DG. Multiple sequence alignments. Current opinion in structural biology. 2005;15(3):261-6.
- Yin X, Jiang XT, Chai B, et al. ARGs-OAP v2. 0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics. 2018;34(13):2263-70.
- Bienert S, Waterhouse A, De Beer TA, et al. The SWISS-MODEL Repository—new features and functionality. Nucleic acids research. 2017;45(D1): D313-9.
- Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic acids research. 2018;46(W1): W296-303.
- Chakraborty AK. A Method of Identification of SARS-CoV-2 Variant Using NCBI BLAST-2 100% Homology Search with Specific Oligonucleotides Selected at the Deletion Boundaries of S, N, ORF7a, ORF8 and ORF1ab Proteins.2022.
- Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of Covid-19 vaccines against the B. 1.617. 2 (Delta) variant. New England Journal of Medicine. 2021;385(7):585-94.
- Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276-80.
- Zhang Y, Chen Y, Li Y, et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proceedings of the National Academy of Sciences. 2021;118(23): e2024202118.
- Wu X, Xia T, Shin WJ, et al. Viral mimicry of interleukin-17A by SARS-CoV-2 ORF8. MBio. 2022;13(2): e00402-22.
- Kee J, Thudium S, Renner DM, et al. SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry. Nature. 2022;610(7931):381-8.
- Pandey P, Prasad K, Prakash A, et al. Insights into the biased activity of dextromethorphan and haloperidol towards SARS-CoV-2 NSP6: in silico binding mechanistic analysis. Journal of Molecular Medicine. 2020; 98:1659-73.
- Ricciardi S, Guarino AM, Giaquinto L, et al. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature. 2022;606(7915):761-8.
- Zhu FC, Guan XH, Li YH, et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. The Lancet. 2020;396(10249):479-88.
- Rajah MM, Hubert M, Bishop E, et al. SARSâ?CoVâ?2 Alpha, Beta, and Delta variants display enhanced Spikeâ?mediated syncytia formation. The EMBO journal. 2021;40(24): e108944.
- Ku Z, Xie X, Davidson E, et al. Molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape. Nature communications. 2021;12(1):469.
- Cottam EM, Maier HJ, Manifava M, et al. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy.2011;7(11):1335-47.
- Pyke AT, Nair N, van den Hurk AF, et al. Replication kinetics of B. 1.351 and B. 1.1. 7 SARS-CoV-2 variants of concern including assessment of a B. 1.1. 7 mutant carrying a defective ORF7a gene. Viruses. 2021;13(6):1087.
- Chakraborty AK. Dynamics of SARS-Cov-2 ORF7a Gene Deletions and Fate of Downstream ORF7b and ORF8 Genes Expression.2022.
- Chakraborty AK. SARS-CoV-2 ORF8 gene CAA= TAA and AAA= TAA Termination Codon Mutations found mostly in B. 1.1. 7 Variants was Independent of Popular L84S Point Mutations. Int J Clin Med Edu Res. 2022;1(6):192-208.
- Telwatte S, Martin HA, Marczak R, et al. Novel RT-ddPCR assays for measuring the levels of subgenomic and genomic SARS-CoV-2 transcripts. Methods. 2022; 201:15-25.
- Chaudhari AM, Singh I, Joshi M, et al. Defective ORF8 dimerization in SARS-CoV-2 delta variant leads to a better adaptive immune response due to abrogation of ORF8-MHC1 interaction. Molecular Diversity. 2022:1-3.